As you learn to use IGB, you'll find it offers many features that make it one of the best tools available for visualization and exploration of genomic data sets.
If you are new to IGB, the following six step Quick Start Guide will help you get started using IGB.
If you have trouble starting IGB, see the Troubleshooting page or feel free to contact us.
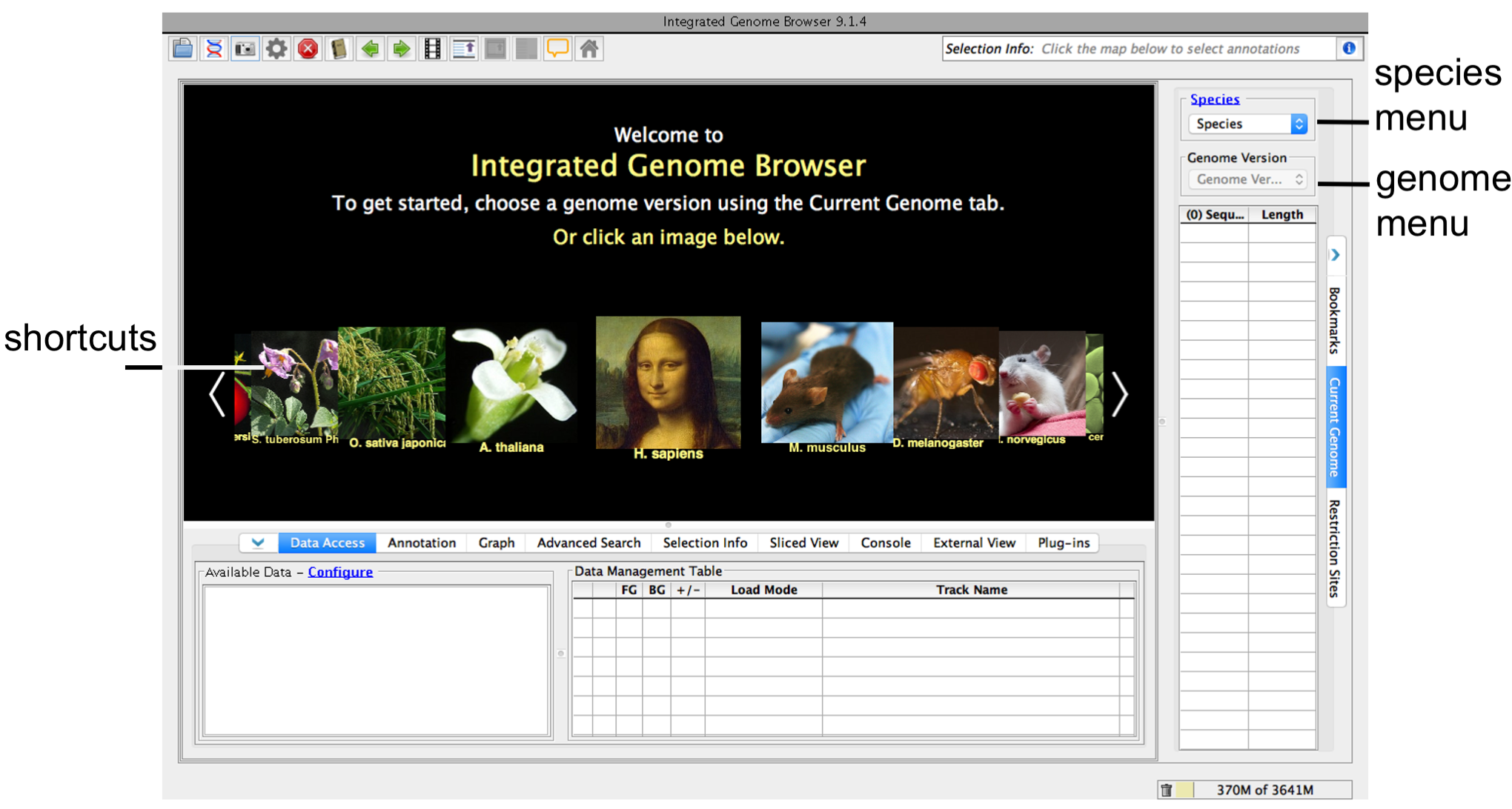
To choose a species and genome version
or
IGB start screen

If your species or genome version is not listed, you can enter it as a custom species and genome when you open a data file.
See also
Open data sets from remote data sources (Data Access tab) or by opening local files.
To open a data set from a remote data source
To open local files on your computer
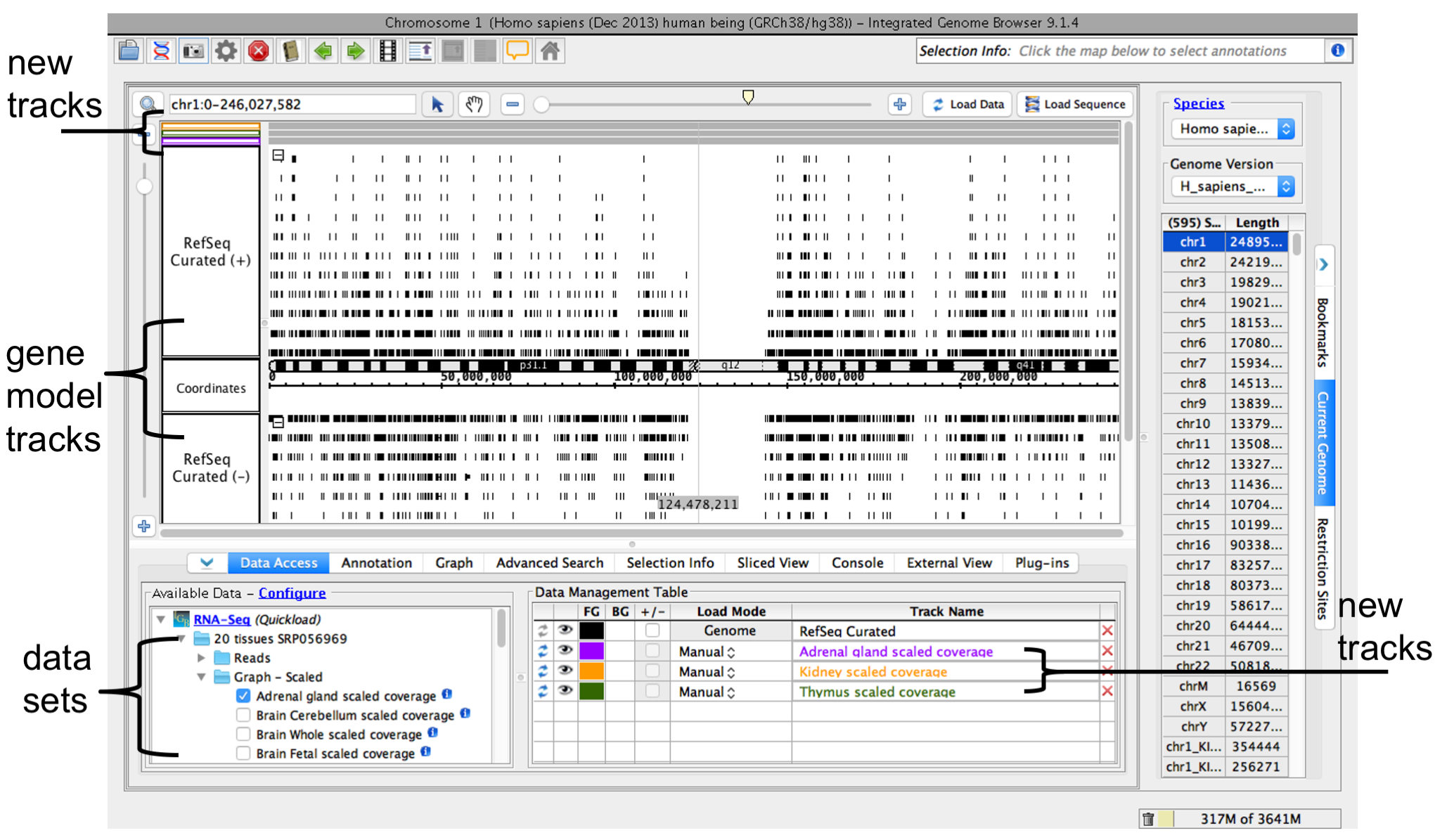
When you select a data sets or file, IGB will add a new empty track to the main view and list it in the Data Management table. Regions that do not have data loaded have a darker background color.
IGB window after opening a data set
If you would like to open a sequence file (fasta or 2bit format) to use as the reference sequence
Because many data files contain too much data to view all at once, IGB does not start loading data into the viewer until you click the Load Data button.
Before loading data, first zoom in to a region you want to view.
To zoom in on a region

Other ways to zoom include
See also:
To load data for the currently visible region, click Load Data button. Regions with loaded data have a lighter background color matching the background color setting for the track.
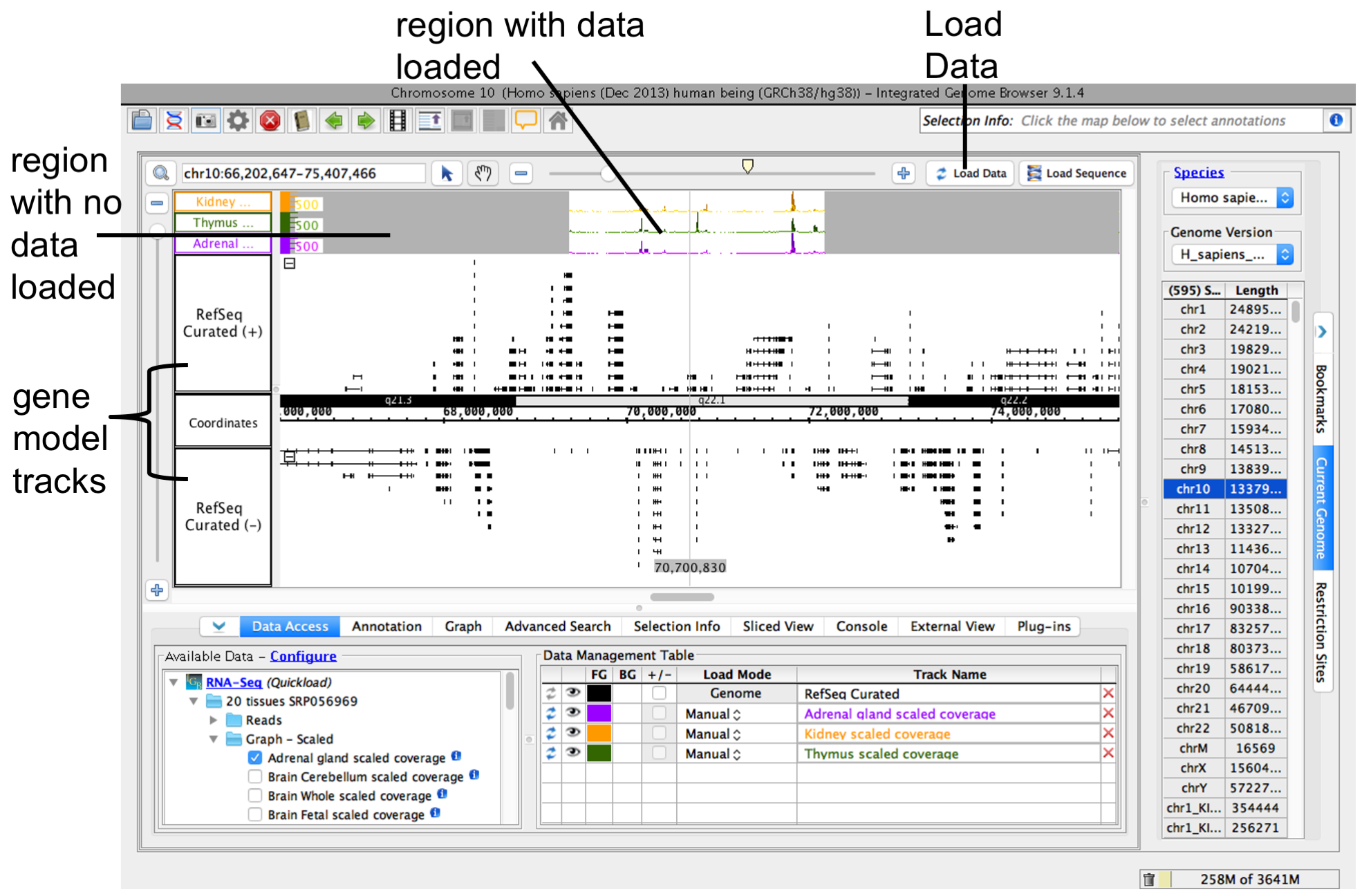
See also:
You can reorder the tracks by dragging the Track Label into the position you want (the Data Management Table reflect changes).
The Annotation and Graph tab will allow you to change color, size, amount of data shown as well as other various functions.
See also: