About IGB QuickLoad
Use IGB QuickLoad (QL) to share data files on-line using a Web site or cloud storage service. Using IGB QuickLoad, you can set up and configure data sets so that they'll be listed in the IGB Data Access panel, making it easy for lab members, collaborators, and members of the public to browse and load your data as new tracks in IGB.
QuickLoad sites can be local or remote. Typically, you'll create a QuickLoad site on your local computer, test it there, and once you're happy with how the data look when loaded into IGB, you'll transfer it to a Web site.
How to set up an IGB QuickLoad site
First, find out if your genome is already supported by the main IGB QuickLoad site.
If your genome is already there, then you should make a satellite QuickLoad site. A satellite site should not contain sequence data or gene models annotations as these are already available from the main IGB QuickLoad site.
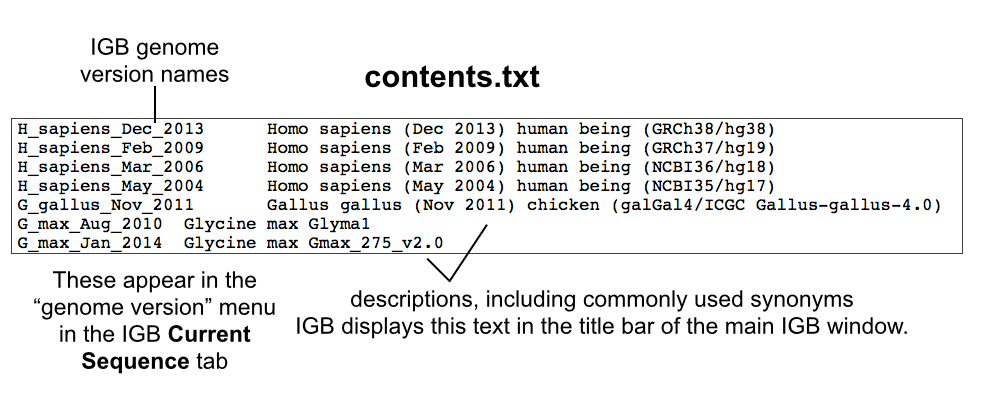
To find out if your genome is already supported, look for it in the the main IGB QuickLoad site's contents.txt file.
If your genome is listed, go to: Create a satellite IGB QuickLoad site for genomes already supported in the main IGB QuickLoad.
If your genome is not listed, go to: Sharing data for a new or custom genome not already part of IGB QuickLoad.