General Function Checklist
Download and unzip test genomes.zip
The folder "test genomes" should contain the following files:
- E_unicornis_Jul_2043.bed.gz.tbi
- E_unicornis_Jul_2043.bed.gz
- E_unicornis_Jul_2043.2bit
- T_rex_Jun_1993.2bit
Start IGB 9.1.8 or later.
Select the Plug-ins tab.
Click Launch App Manager.
In the IGB App Manager window:
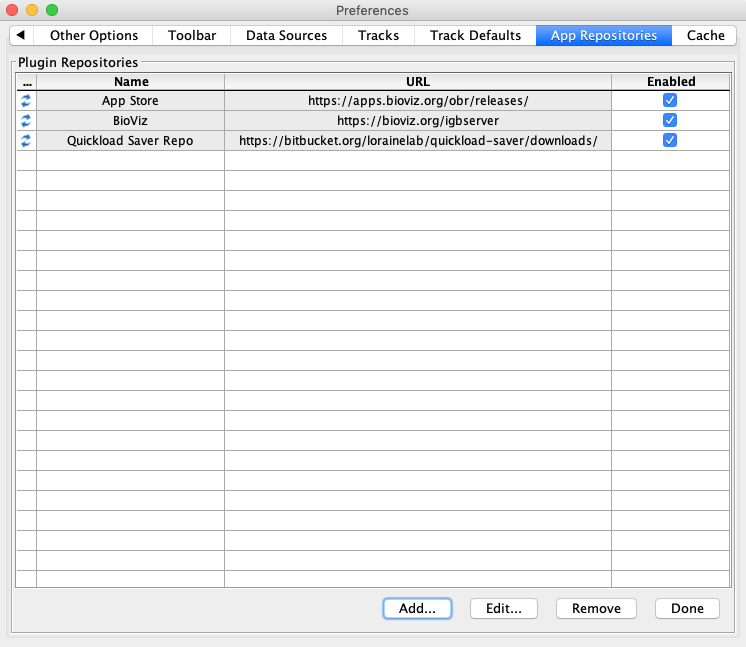
Select Manage Repositories...
In the Preferences window App Repositories tab:
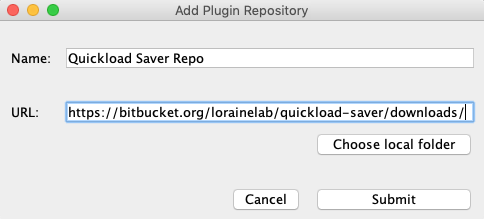
Click Add...
In the Add Plugin Repository window enter the following information:
- Name: Quickload Saver Repo
Click Submit
In the Preferences window App Repositories tab:
Click Done
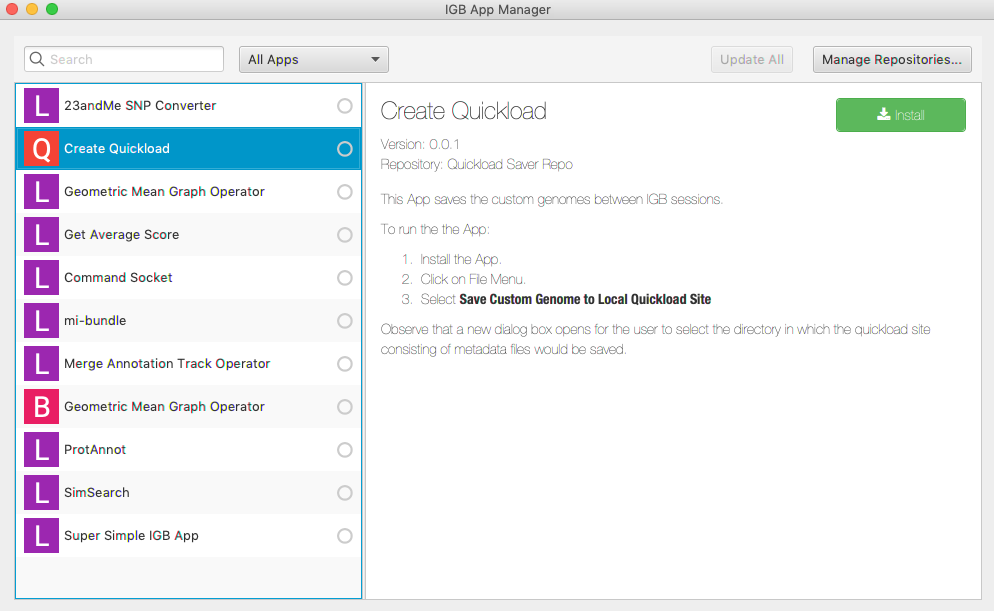
In the IGB App Manager window:
Select the Create Quickload app.
Click Install.
Checkpoint:
- mac
- linux
- windows
Close the IGB App Manager window.
In IGB:
Select File > Open Genome from File...
UPDATE AFTER 2783 MERGE
Select E_unicornis_Jul_2043.2bit
ENTER THE INFORMATION IN THE OPEN GENOME FROM FILE WINDOW AND CLICK OK
Select File > Open File...
Select E_unicornis_Jul_2043.bed.gz
Click Open
In the Data Access tab in the Data Management Table section:
Change the Load Mode to Genome for E_unicornis_Jul_2043.bed.gz
Change the color of the foreground (FG) to red
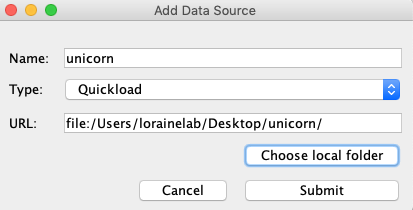
Select File > Save Custom Genome to Local Quickload Site
Select your desktop as the destination and unicorn as the name.
In IGB:
Select File > Open Genome from File...
UPDATE AFTER 2783 MERGE
Select T_rex_Jun_1993.2bit
ENTER THE INFORMATION IN THE OPEN GENOME FROM FILE WINDOW AND CLICK OK
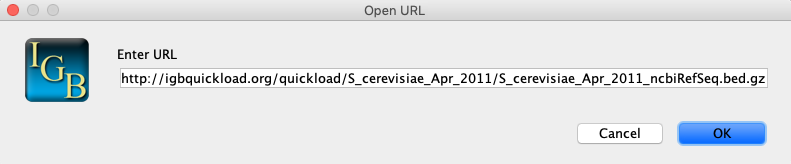
Select File > Open URL...
In the Open URL window:
Enter URL: http://igbquickload.org/quickload/S_cerevisiae_Apr_2011/S_cerevisiae_Apr_2011_ncbiRefSeq.bed.gz
Click OK
In IGB:
In the Data Access tab in the Data Management Table section:
Change the Load Mode to Genome for S_cerevisiae_Apr_2011_ncbiRefSeq.bed.gz
Change the color of the foreground (FG) to green.
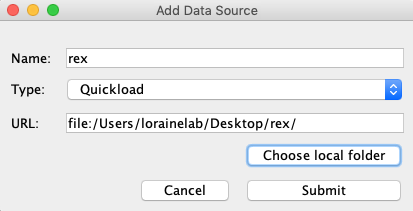
Select File > Save Custom Genome to Local Quickload Site
Select your desktop as the destination and rex as the name.
In IGB:
Select File > Preferences...
In the Preferences window select the Other Options tab.
Click Reset Preference to Defaults
Open IGB
In IGB:
Select File > Preferences
In the Preferences window:
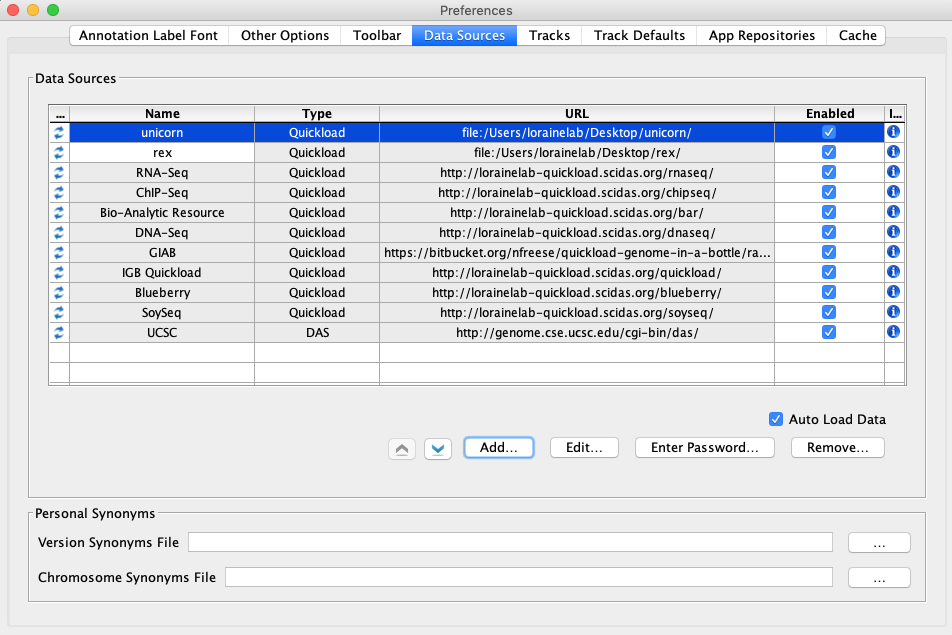
Select the Data Sources tab.
In the Data Sources tab:
Click Add...
In the Add Data Source window:
- Name: unicorn
- Type: Quickload
- Click Choose local folder
Select the unicorn folder you created earlier and click Open
In the Add Data Source tab:
Click Submit
In the Data Sources tab:
Click Add...
In the Add Data Source window:
- Name: rex
- Type: Quickload
- Click Choose local folder
Select the rex folder you created earlier and click Open
In the Add Data Source tab:
Click Submit
Checkpoint:
- mac
- linux
- windows
Close the Preferences window
In IGB:
Select the Log tab and click Clear
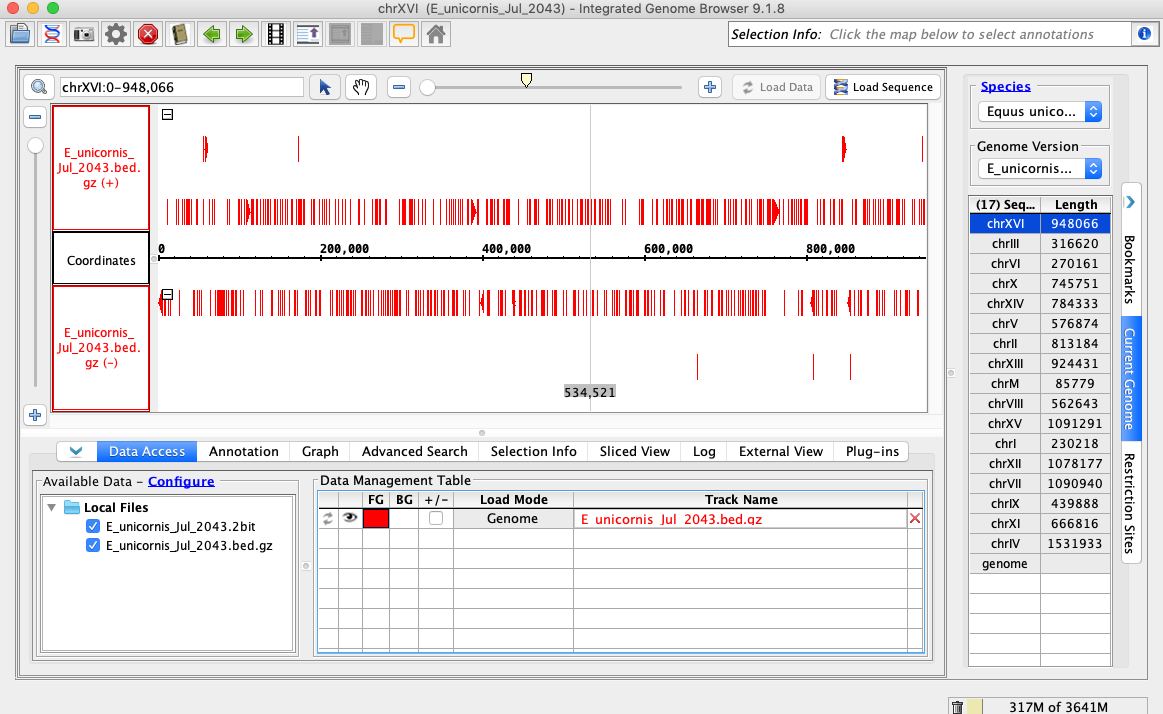
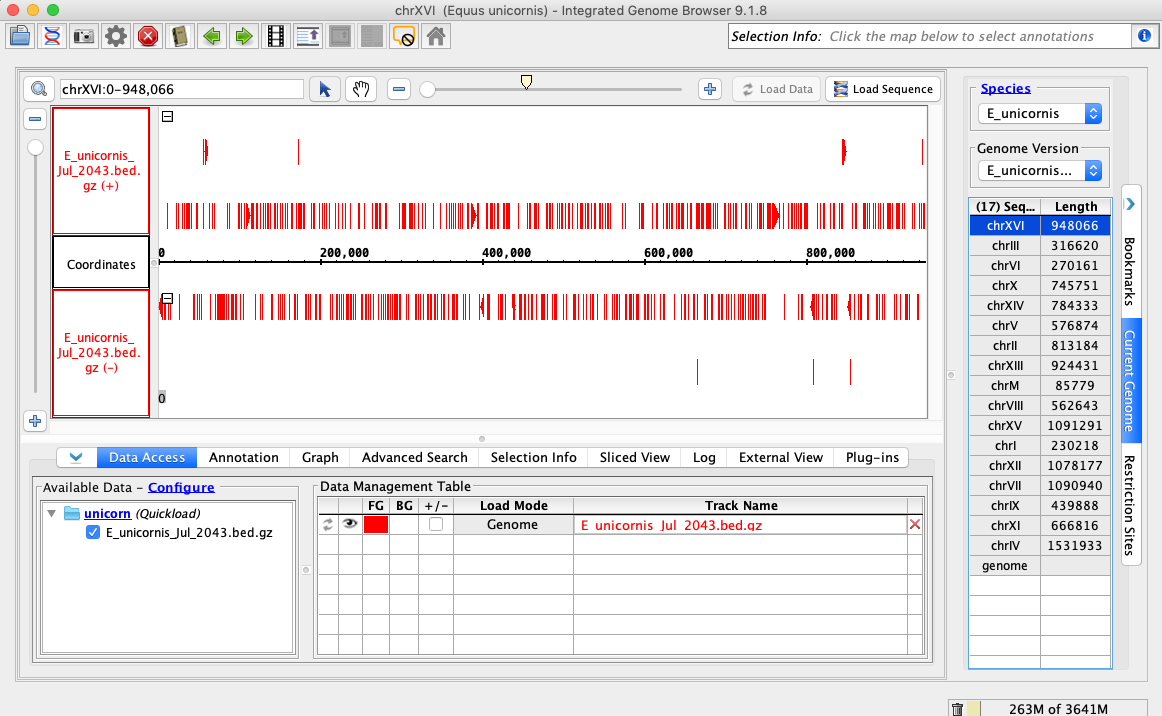
In the Species dropdown select E_unicornis
In the Genome Version dropdown select E_unicornis_Jul_2043
In the Data Access tab, E_unicornis_Jul_2043.bed.gz should be set to Load Mode Genome and should be red (FG)
Compare your IGB to the picture and check the box if they appear to be the same:
- mac
- linux
- windows
Navigate to chrXVI:2,618-2,661
Click Load Sequence
SHOULD LOAD SEQUENCE, NOT WORKING?
Select the Log tab and check for errors.
Click Clear in the Lob tab if there are no errors.
Checkpoint:
- mac
- linux
- windows
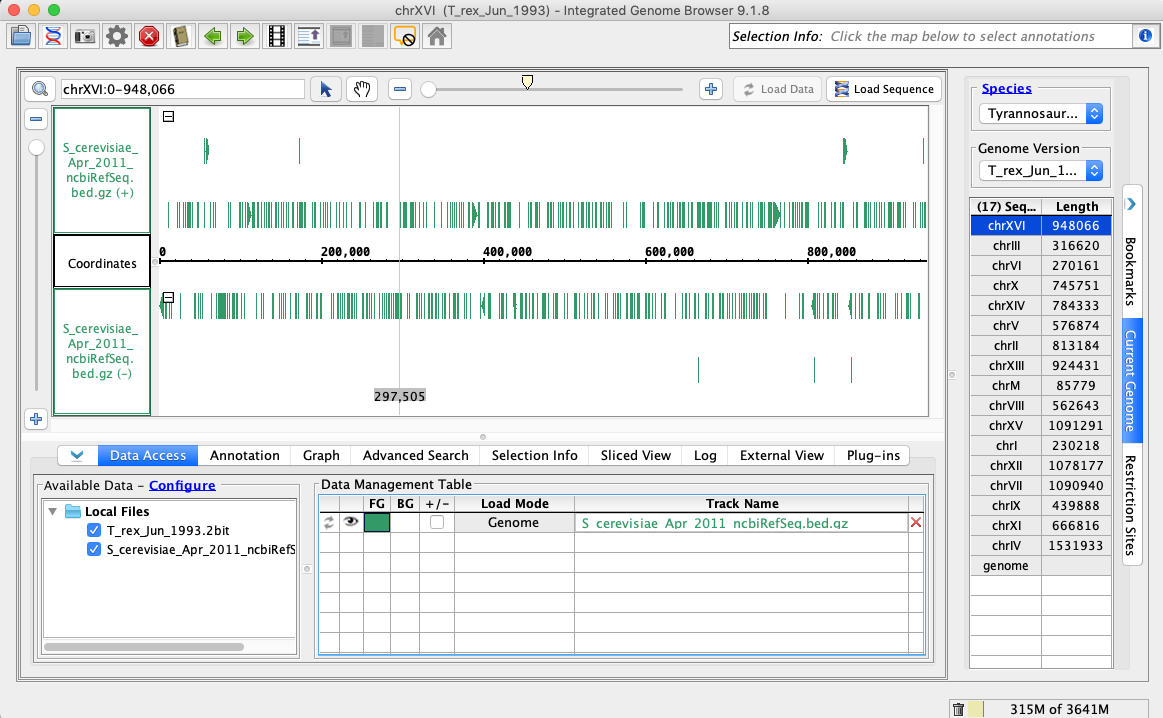
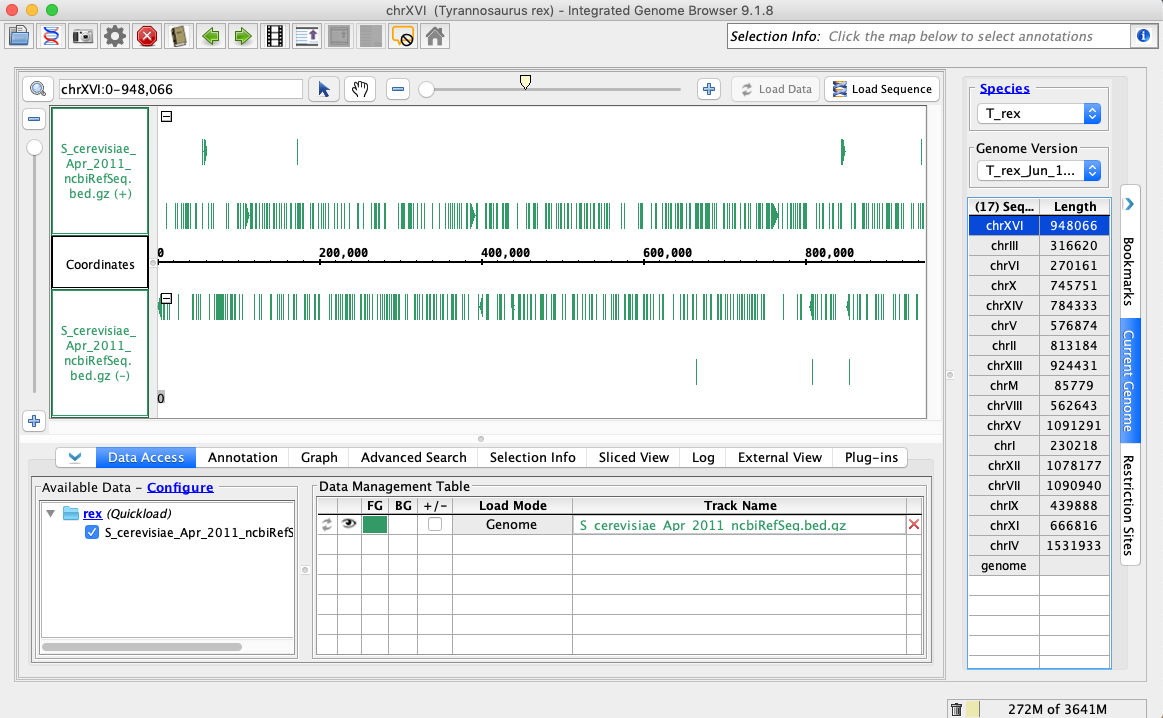
In the Species dropdown select T_rex
In the Genome Version dropdown select T_rex_Jun_1993
In the Data Access tab, S_cerevisiae_Apr_2011_ncbiRefSeq.bed.gz should be set to Load Mode Genome and should be green (FG).
Compare your IGB to the picture and check the box if they appear to be the same:
- mac
- linux
- windows
Navigate to chrXVI:2,618-2,661
Click Load Sequence
SHOULD LOAD SEQUENCE, NOT WORKING?
Select the Log tab and check for errors.
Click Clear in the Log tab if there are no errors.
Checkpoint:
- mac
- linux
- windows