Skip to end of metadata
Go to start of metadata
You are viewing an old version of this page. View the current version.
Compare with Current
View Page History
« Previous
Version 18
Next »
General Function Checklist
- Protannot can be launched from IGB
- Go to any genome
- Select one or more whole gene model annotation
- Select the Tools Menu
- Select Start ProtAnnot
- Checkpoint
- sr45a should look as follows when loaded into ProtAnnot. Take note of key items in red.

- The ProtAnnot window title should contain the absolute sequence coordinates, strand direction, contig name, and the genome version.

- InterProScan
- Select ProtAnnot menu
- Select Load InterProScan... menu item
- Verify the InterProScan Job Configuration window appears similar to the image below
- Verify tool tips in the information icon wrap appropriately
- Verify the link in the highlighted box below links to the appropriate InterProScan about page
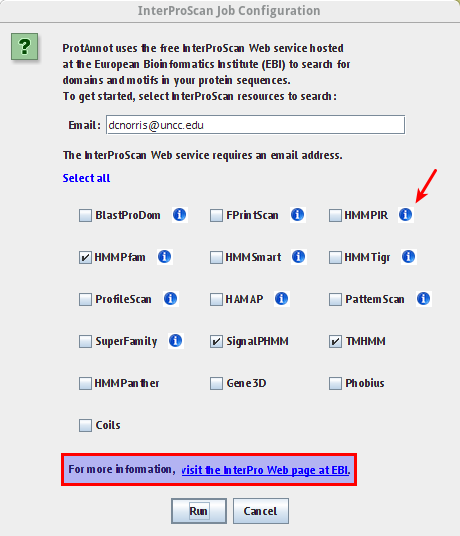
- Select run
- {{ image of InterProScan results }}
- Verify that the Exons have the correct protein sequences

- Select an InterProScan result and verify that the properties are correct
- Correct start, end, and length.
- Functioning URL when clicked
- Application and library name are present

- Open the InterProScan tab
- Verify that that both jobs have a status of FINISHED
- Clicking on the result cell should open the result xml on the InterProScan website







