Introduction
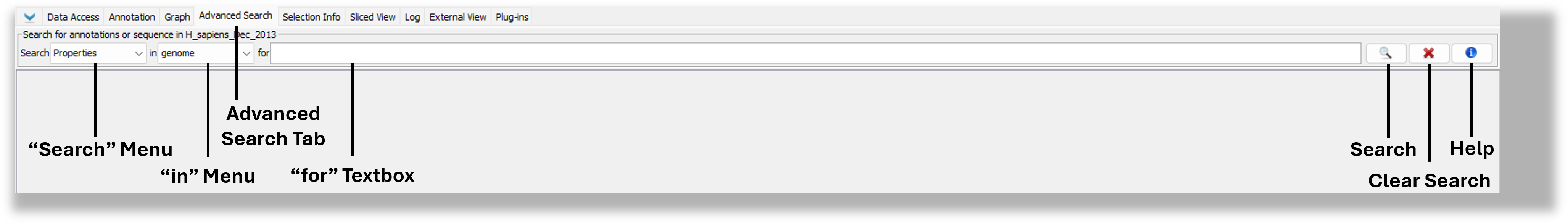
Use the Advanced Search tab to search for gene annotations or sequence residues. Both search types support regular expressions and wild card characters (see the "Regular expression, wild cards, and nucleotide symbols" section below).
Using Advanced Search, you can:
- Look up genes or other annotations by name or keyword
- Find instances of transcription factor binding sites
- Display locations of PCR primers
Search results will appear in the Advanced Search tab in a results table. Double-click a row in the table to view the result in the main IGB window.
If you search for sequence residues, IGB will also display color-coded bars in the coordinates track indicating the matched sequence.
Search types
The Advanced Search supports:
- ID, Name, or Title - find annotations by name
- Keyword - find annotations by keyword
- Residues - find sequences or regular expressions
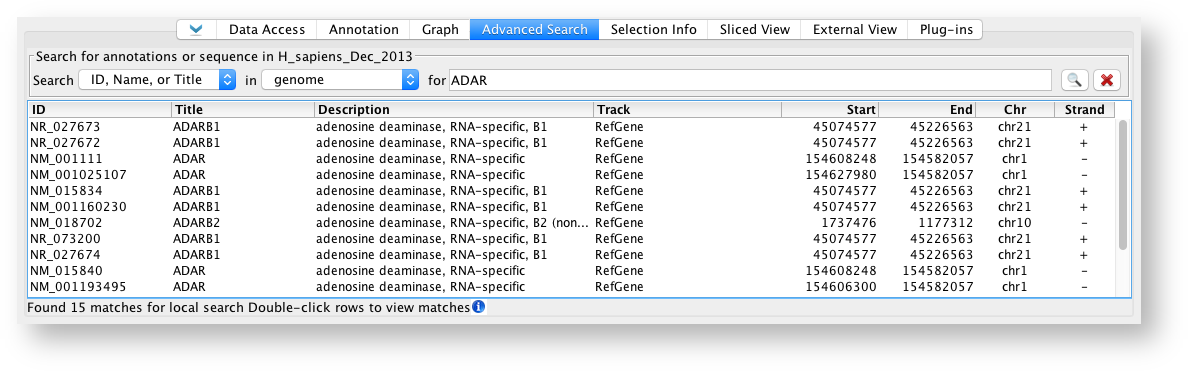
Search by ID, Name, or Title
ID, Name, or Title search will search IDs and names of annotations.
To find an annotation by ID, Name, or Title:
- Select ID, Name, or Title from the Search menu.
- Choose "genome" or a specific chromosome from the in menu.
- Enter the ID or name of the annotation you want to find (for textbox).
- Press <Enter> key or click the Search button.
Only data already loaded into the IGB viewer will be searched.
ID, Name, or Title Search Results
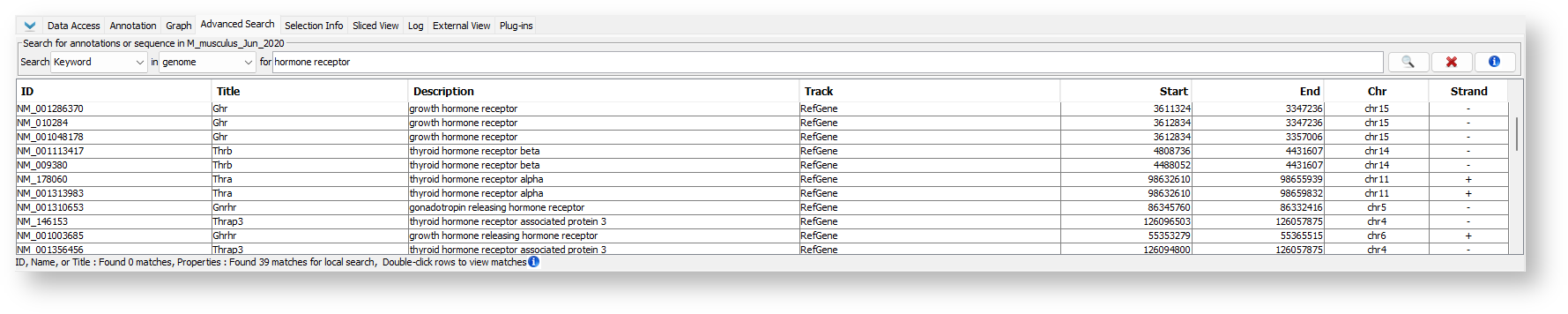
Search by Keyword
Keyword search, similar to ID, Name, or Title search, will search annotation IDs, but it will also search other information associated with annotations such as descriptions and other attributes.
To find an annotation by Keyword:
- Select Keyword from the Search menu.
- Choose "genome" or a specific chromosome from the in menu.
- Enter the keyword you want to search for (for textbox).
- Press <Enter> key or click the Search button.
Keyword Search Results
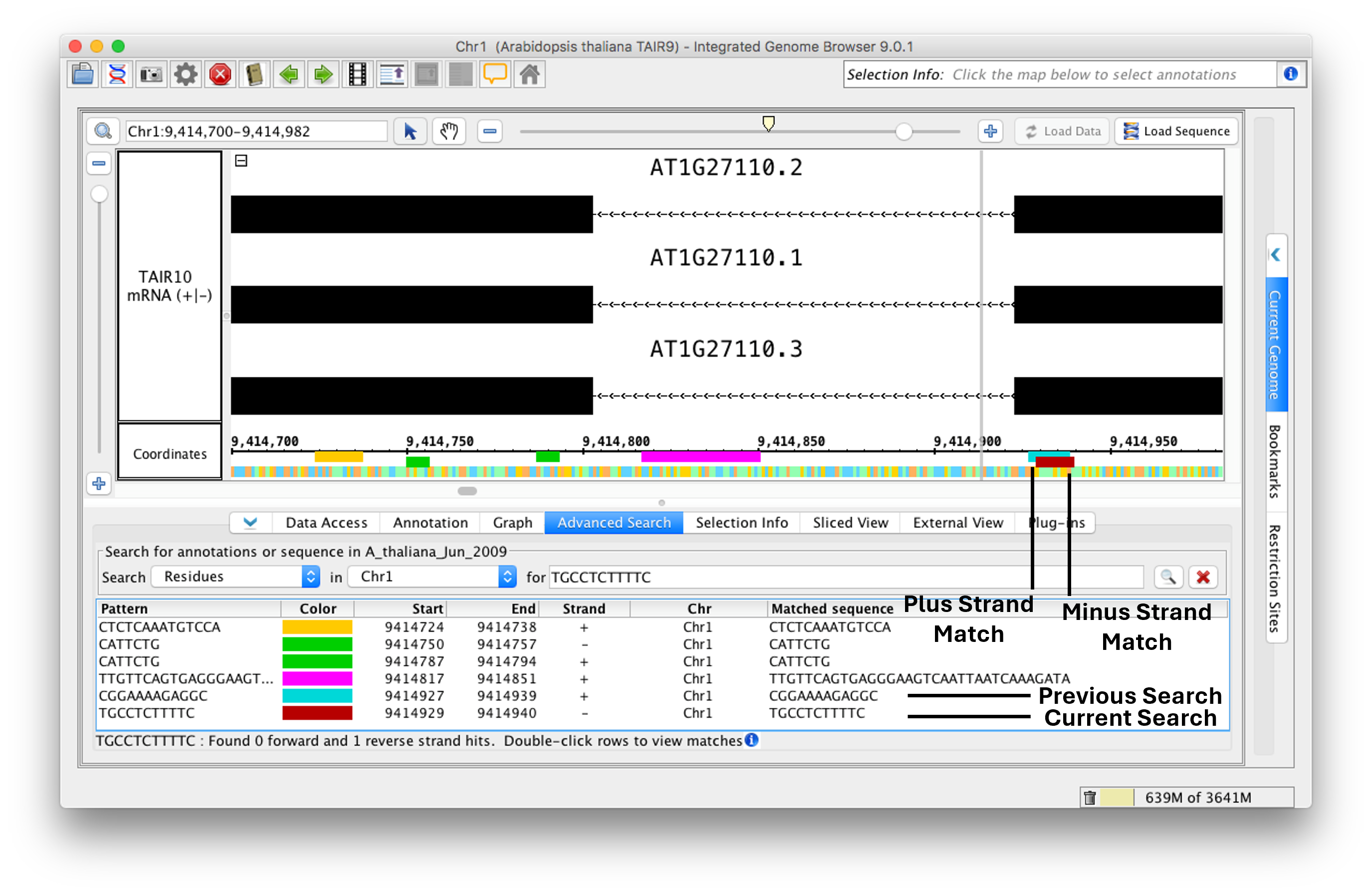
Search by Residues
To find all instances of a sequence or regular expression:
- Select Residues from the Search menu
- Choose "genome" or a specific chromosome from the in menu.
- Enter the sequence or regular expression you want to find (for textbox).
- Press <Enter> key or click the Search button.
- Enter new search terms. Notice that IGB will overlay results, preserving results from previous searches.
IGB displays matches in the results table and as colored bars underneath the coordinates axis. Matches on the minus strand appear in a slightly lower position.
Residues Search Results
Regular expression, wild cards, and nucleotide symbols
IGB searching supports regular expressions and wild cards. This is especially useful when searching for sequence motifs, such as transcription factor binding sites.
Searching by nucleotide symbols is available in IGB versions 9.1.12 and above.
Example queries:
Pattern | Represents | Example | Finds |
. | Any single nucleotide | ACCT.T | ACCTTT, ACCTAT, ACCTGT, and ACCTCT (4 possibilities) |
.. | Any two nucleotides | ACCT..T | ACCTAAT, ACCTATT, ACTAGT, Etc. (4 x 4 possibilities) |
[CG] | C or G | ACCT[CG]TC | ACCTCTC and ACCTGTC |
| X|Y | X or Y | ATC|AAG | ATC and AAG |
T{1,n} | 1 to n T's | ACGGT{1,3}C | ACGGTC, ACGGTTC, ACGGTTTC |
T* | Zero or more T's | ACGGT*C | ACGGC, ACGGTC, ACGGTTC, ACGGTTTC, ACGGTTTTTTTTTTTTTTTTTTTTTTTTTTTC, Etc. |
.*? | A string of any length containing any nucleotides | TCGGGGTTAA.*?CTGGACTC | Many possibilities. Because this allows for so many possibilities, it only recommended with a limited scope of search and/or with very specific (several specified base pairs) on both ends. |
.* | The longest possible string of any length containing any nucleotides | TCGGGGTTAA.*CTGGACTC | Differs from the search above in that the longest possible result(s) will be found. Bear in mind that the result returned from this search with depend on the scope of the search, i.e., how much of the genomic sequence has been loaded and is available for searching. |
| R | A or G | GCCR | GCCA, GCCG |
| Y | C or T | AGCY | AGCC, AGCT |
| S | G or C | AGCS | AGCG, AGCC |
| W | A or T | AGCW | AGCA, AGCT |
| K | G or T | AGCK | AGCG, AGCT |
| M | A or C | AGCM | AGCA, AGCC |
| B | C or G or T | AGCB | AGCC, AGCG, AGCT |
| D | A or G or T | AGCD | AGCA, AGCG, AGCT |
| H | A or C or T | AGCH | AGCA, AGCC, AGCT |
| V | A or C or G | AGCV | AGCA, AGCC, AGCG |
| N | Any base (i.e., A or G or T or C) | AGCN | AGCA, AGCG, AGCT, AGCC |
| \QN\E | N | AGC\QNNN\E | AGCNNN |
More information about regular expressions is available from this Java Regex Cheat Sheet.