General Function Checklist
Objective: Validate that soft-clipping can be hidden and customized using the right-click menu of a bam track, as shown below:
Add the smoke test Data Provider to IGB using the following URL: https://quickload-testing.s3.amazonaws.com/smokeTestingQuickload/
- Open: Homo sapiens genome
- Select: Bam - PacBio with softclipping
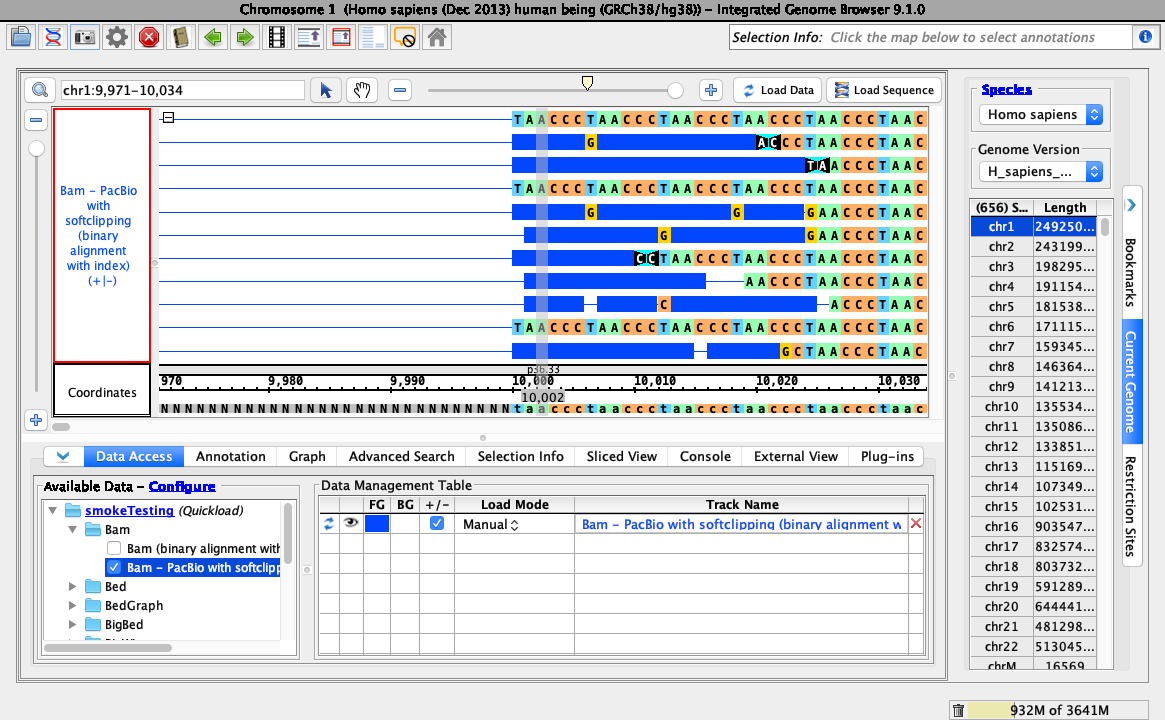
- Navigate to: chr1:9,971-10,034
- Click Load Sequence to load sequence data, click Load Data to load data into it.
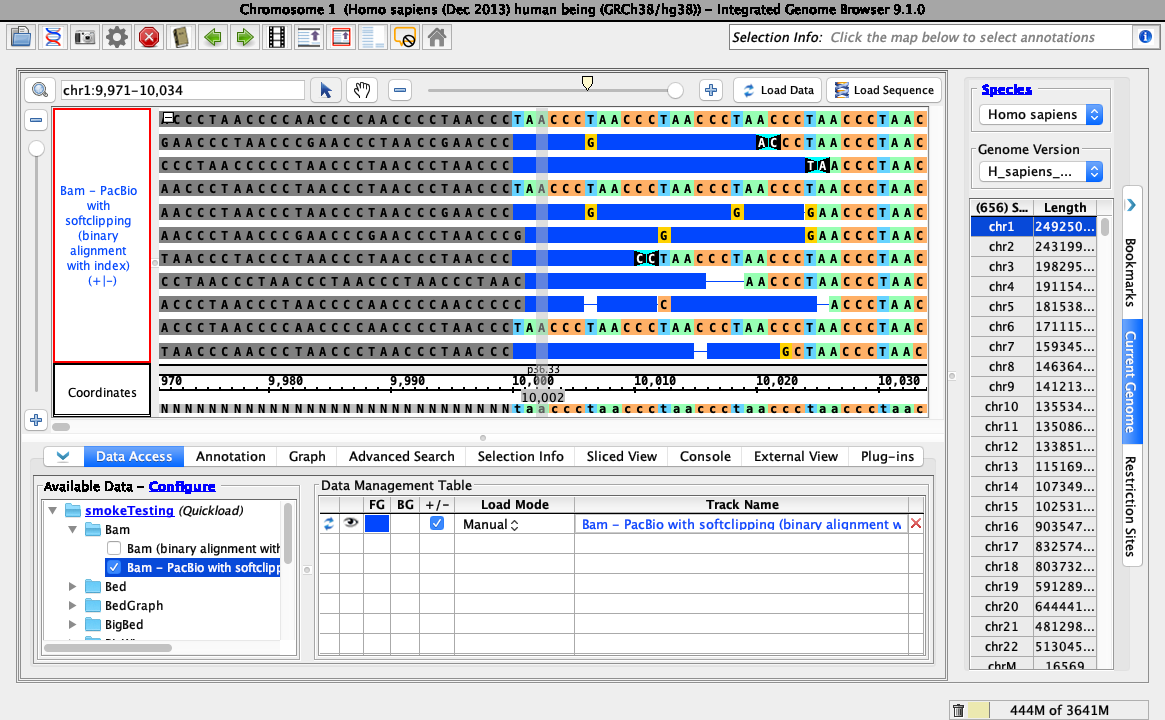
Ensure that loading data looks like this (stack height set at 10, reads may be organized randomly):
- mac
- linux
- windows
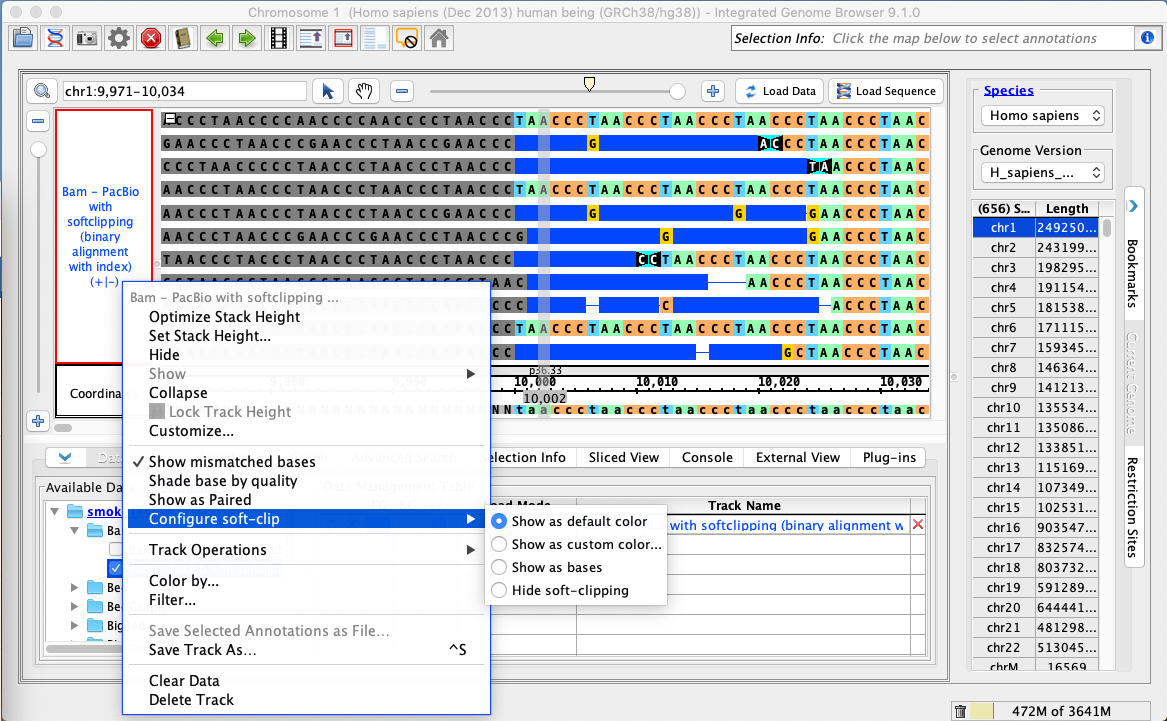
Validate that the 'Configure soft-clip' submenu of the bam track right-click menu has 'Show as default color' selected.
- mac
- linux
- windows
Right-click on the bam track and select:
Configure soft-clip -> Show as custom color
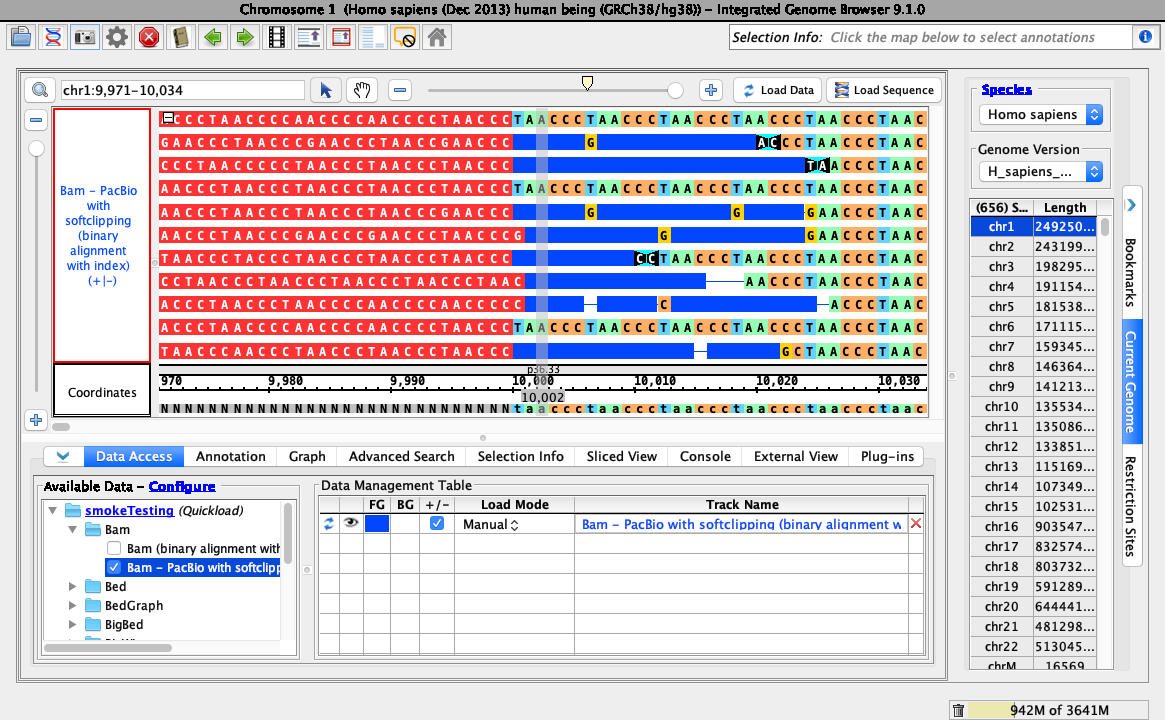
A color picker should appear. Select a color (the example below selected red).
Ensure your view is as below, with the color you chose correctly applied:
- mac
- linux
- windows
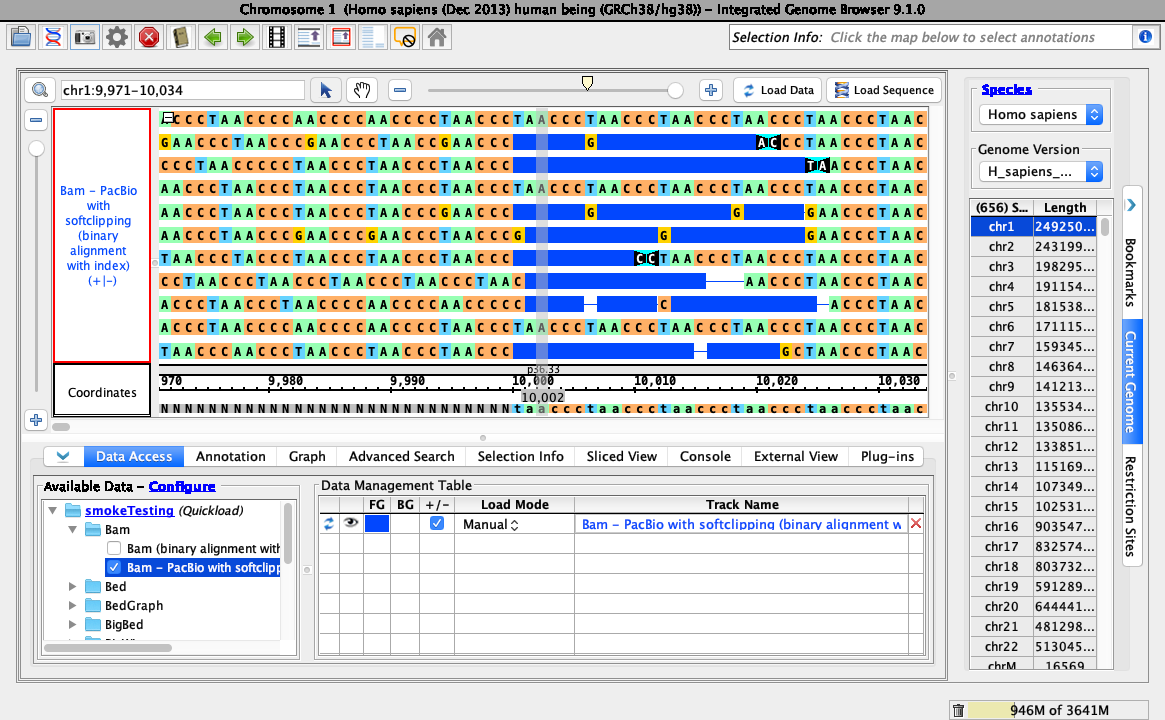
Right-click on the bam track and select:
Configure soft-clip -> Show as bases
Ensure your view is as below:
- mac
- linux
- windows
Right-click on the bam track and select:
Configure soft-clip -> Hide softclipping
Ensure your view is as below:
- mac
- linux
- windows
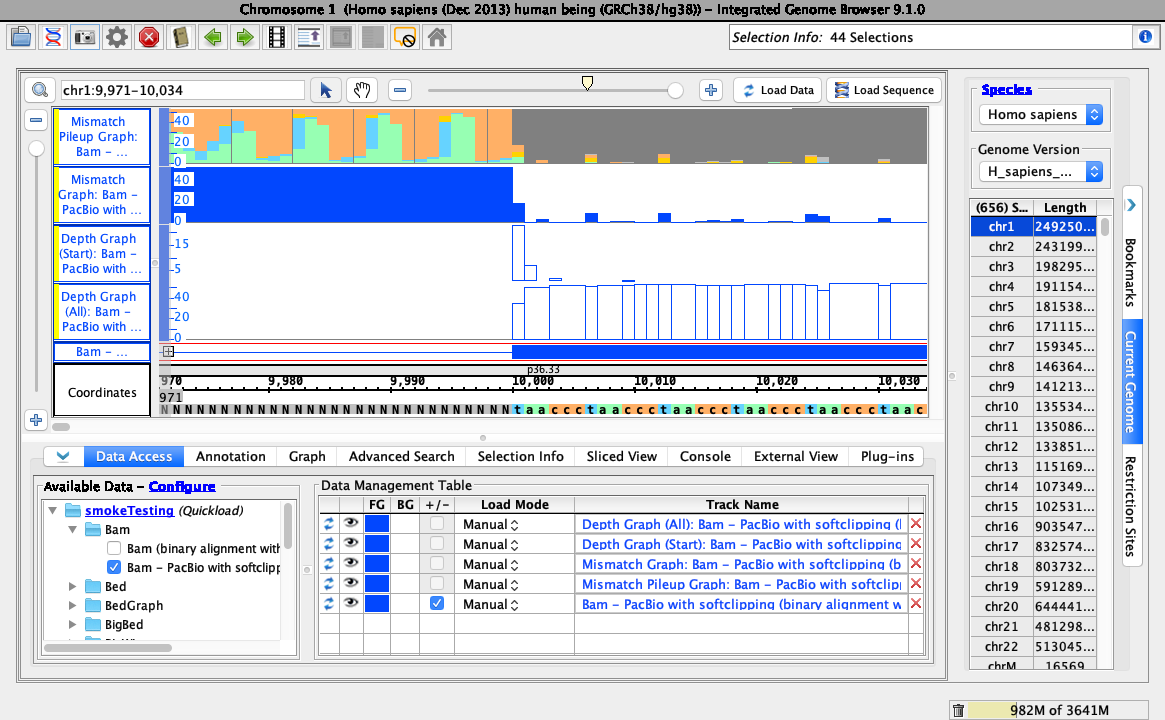
To test track operations are working correctly for soft-clipped data, right-click on the bam track and select:
- Track Operations > Depth Graph (All)
- Track Operations > Depth Graph (Start)
- Track Operations > Mismatch Graph
- Track Operations > Mismatch Pileup Graph
- Collapse
Ensure your view is as below:
- mac
- linux
- windows