...
- Open the human genome (H_sapiens_Dec_2013) in IGB.
- Navigate to: chr1:3,320-26,768
- Hide the RefSeq Curated annotation track by right-clicking and selecting Hide.
- Select File > Open URL...Enter: https://quickload-testing.s3.amazonaws.com/smokeTestingQuickload/H_sapiens_Dec_2013/Bam/Download the two files below:
- In IGB, select File > Open File...
- Select the pacBio.bam file and click Open.bai
- Click OK
- Click Load Sequence to load sequence data.
- Click Load Data to load data into it.
Soft-clipping is enabled by default in IGB, with the soft-clipped section of the read displayed in gray.
...
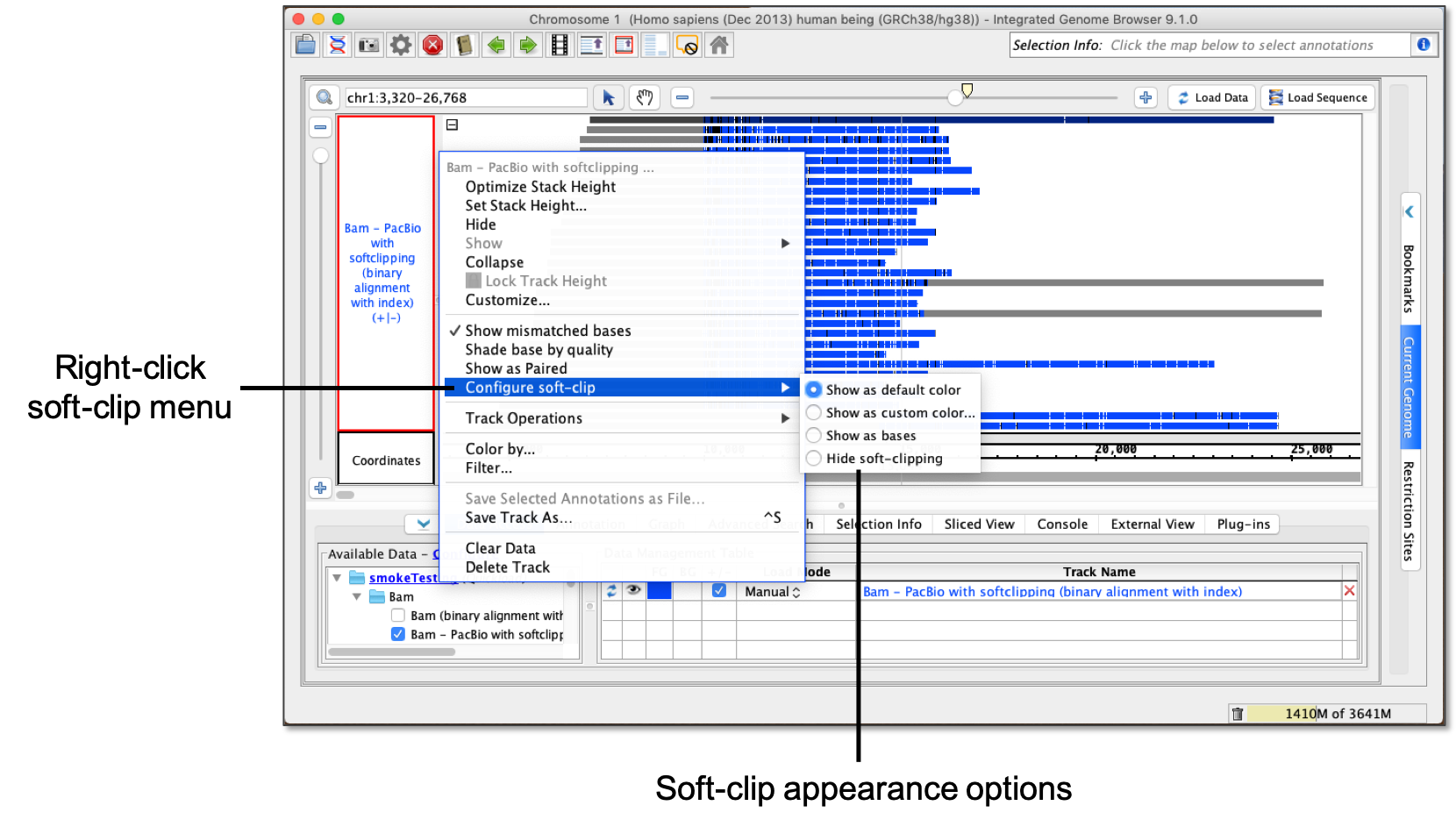
- Right-click the reads track label.
- Select Configure soft-clip.
Options for viewing soft-clipping include:
- Show as default color - Soft-clipping shows as a default gray.
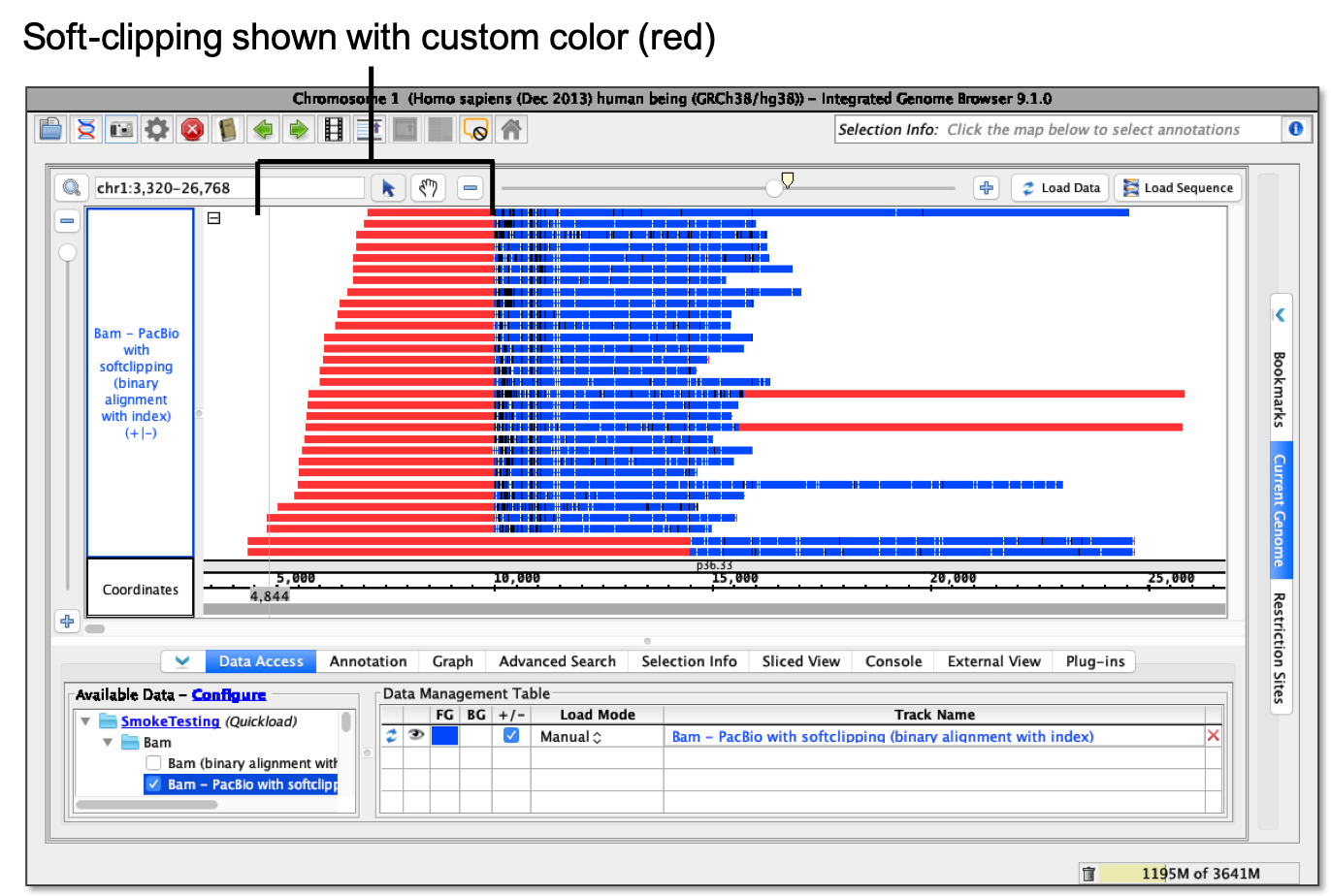
- Show as custom color... - Using a color picker, soft-clipping shows as selected color.
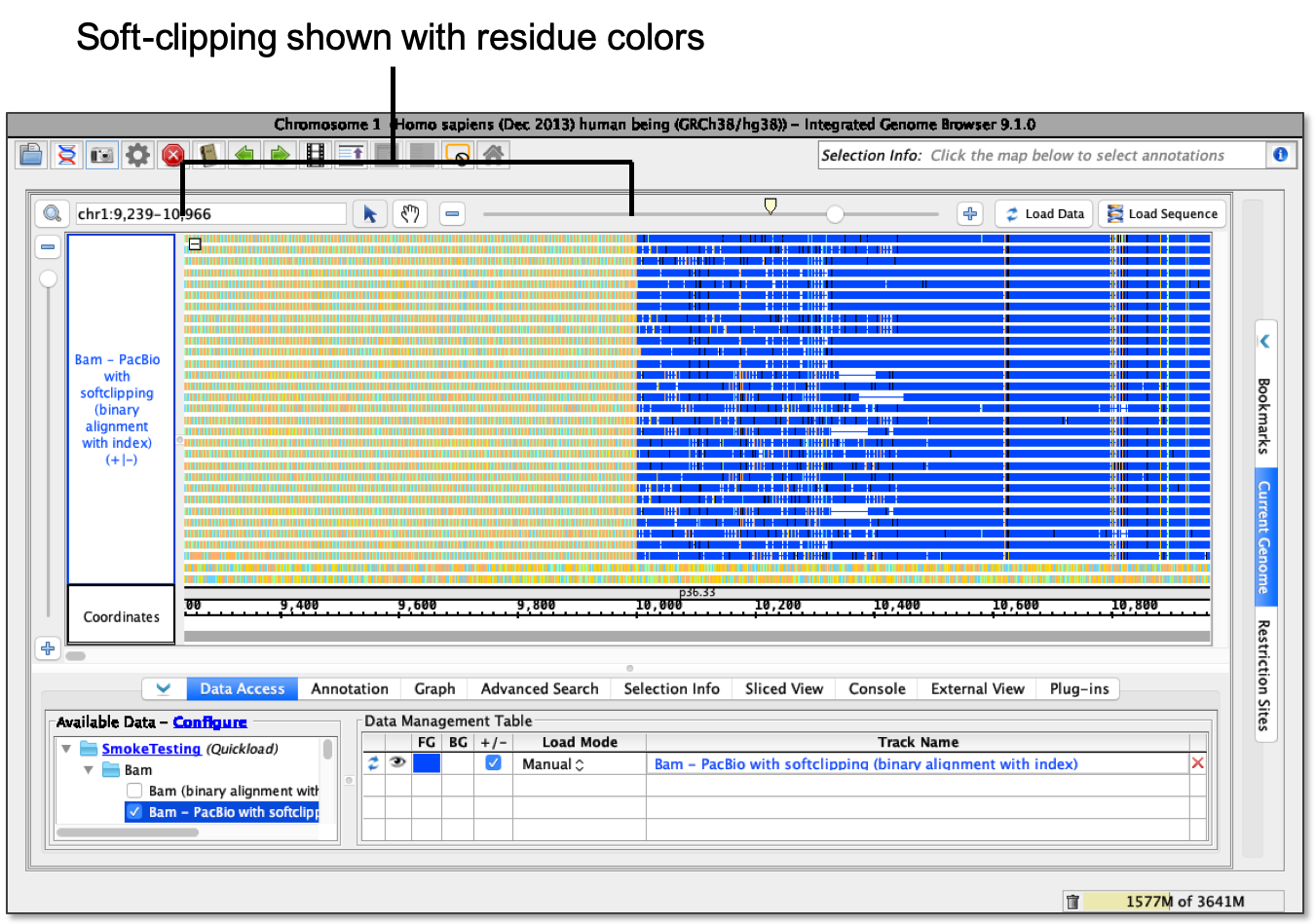
- Show as bases - Soft-clipping shows as Residue Colors.
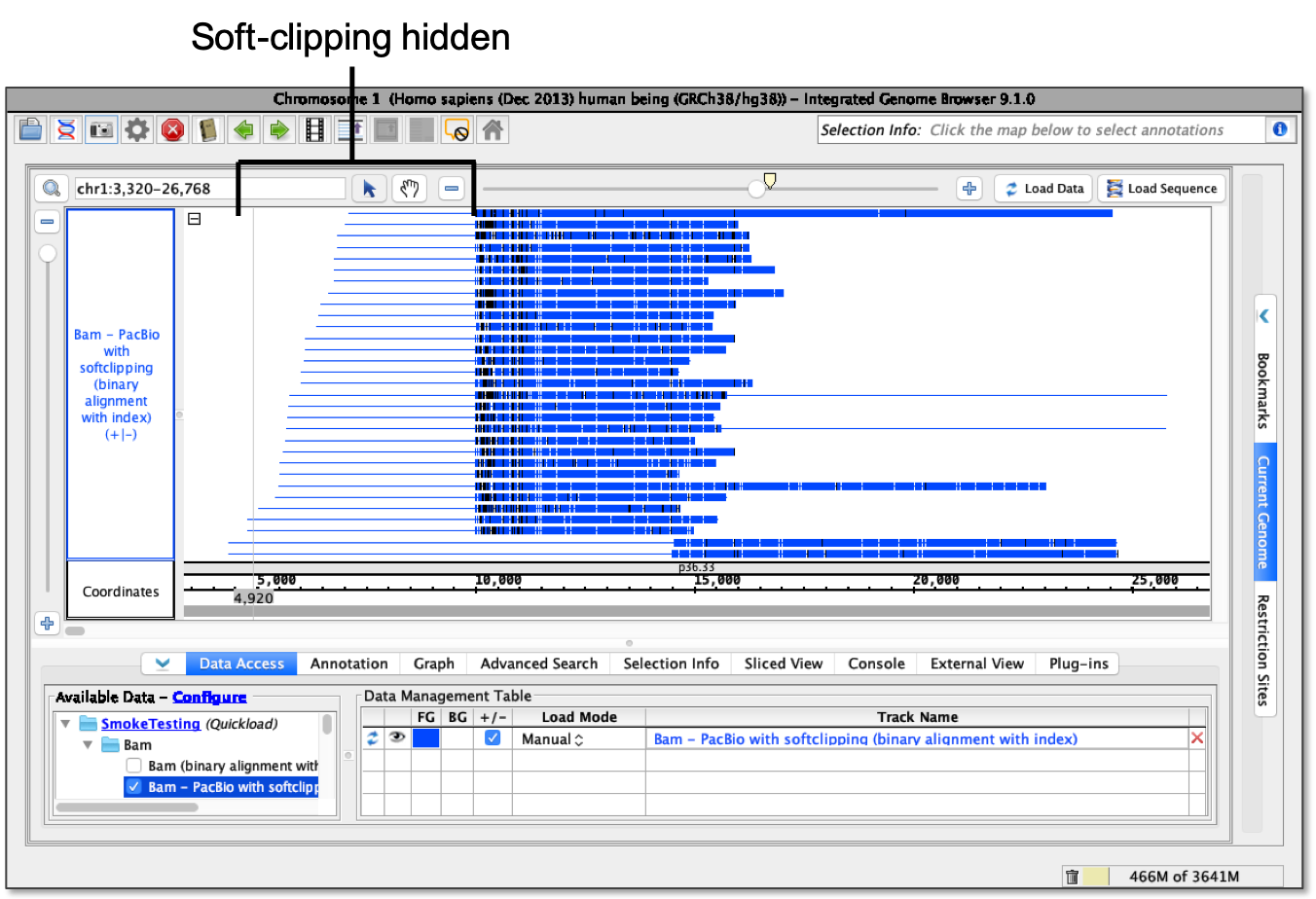
- Hide soft-clipping - Soft-clipping is hidden.
Note: When using the hide soft-clipping option, a line showing the full length of the read including the soft-clipped portion will appear.
...
- Depth Graph (All) and Depth Graph (Start) ignore the soft-clipped parts of reads.
- Mismatch Graph and Mismatch Pileup Graph include the soft-clipped parts of reads.
Data used in these examples is a subset taken from the Genome in a Bottle consortium (HG002 PacBio CCS 10kb):
Zook JM, Catoe D, McDaniel J, et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3:160025. Published 2016 Jun 7. doi:10.1038/sdata.2016.25
...