General Function Checklist
- Loading Default Tracks
...
Validating Track Preference Changes
- Open
...
- Zoom,Drag, and Selection Operations
...
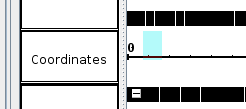
- Zoom in by click and dragging across the coordinates. Note that the screenshot is missing the mouse cursor. The blue highlighted area represents the area created by dragging the cursor.
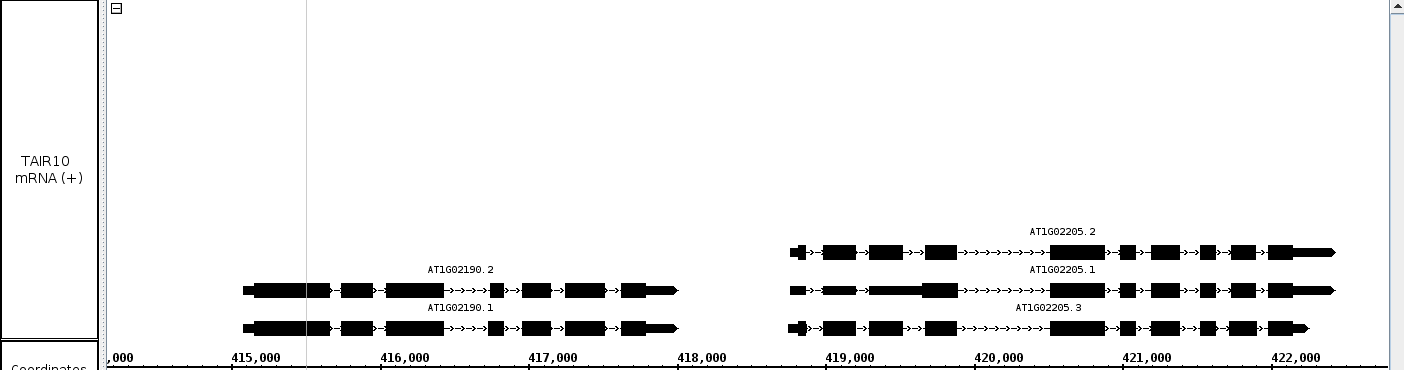
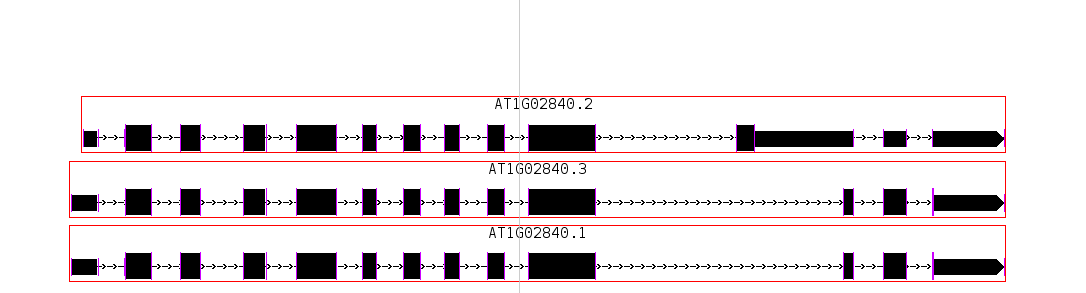
- Adjust the zoom such that multiple gene models are in view
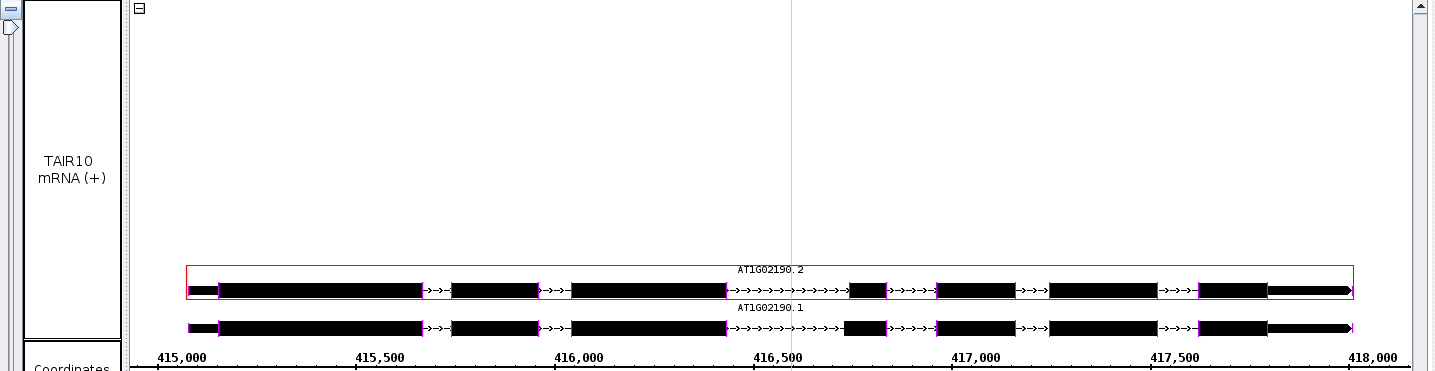
- Double click on the barb wire region of a gene model. The view should zoom to fit the selected gene model.
- Zoom back out to view multiple gene models
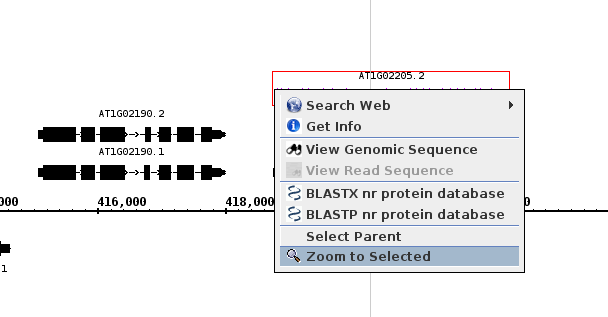
- Right click on a different gene model.
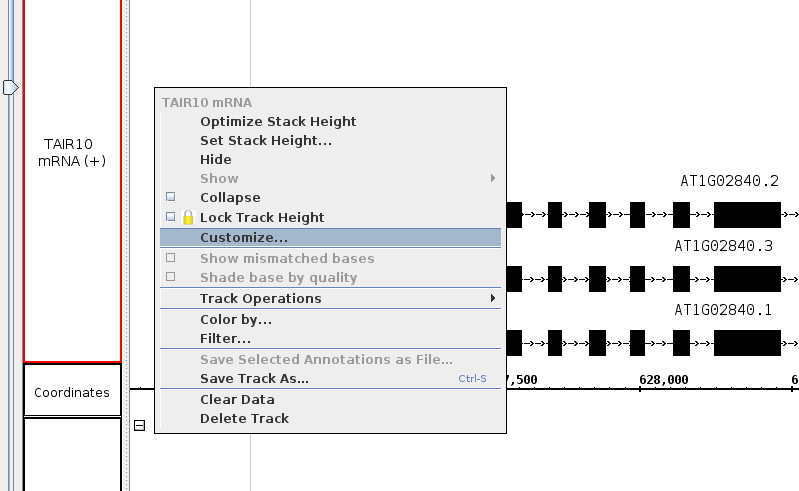
- Validate that the menu looks as follows
- Select "Zoom to Selected"
- The view should zoom to fit the selected gene model
- Zoom slightly out in both the x and y direction.
- Select the hand "Grab Tool".
- Click and drag anywhere in the track view.
- Validate that the panel scrolls in the direction of the cursor.
- Select the "Selection Tool"
- Go to a group of gene models that are stacked
- Click and drag across the gene models.
- Validate that all of the gene models are selected.
- Track Style Preferences
...
- the A_thaliana_Jun_2009 genome.
- Navigate to the following coordinates: Chr1:635,037-637,360
- Right click on the track label and select Optimize Stack Height.
- Right click on the white space within a track.
- Select
...
- Customize...
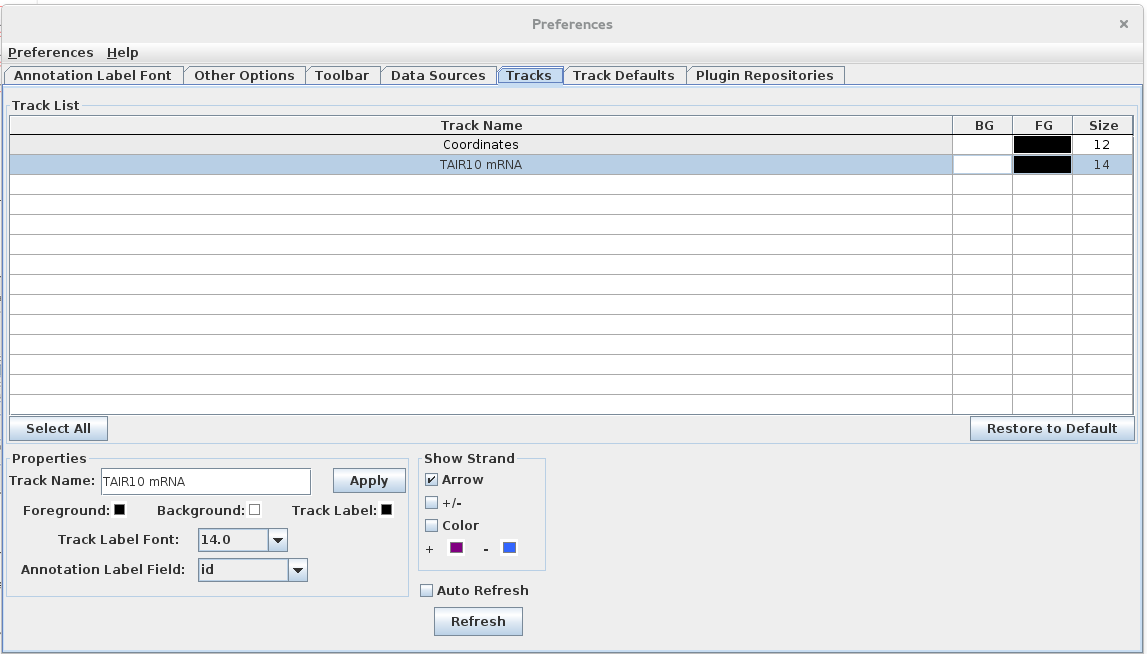
- This action should open opened the Track Preferences tab
...
- with the selected track highlighted.
- mac
- linux
- windows
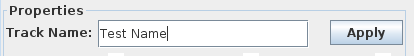
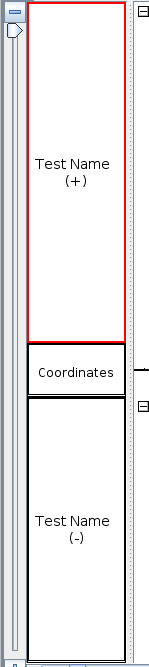
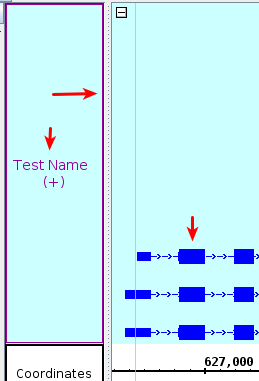
Change the Track Name
...
to Test Name, then select Apply.
...
- The track names were updated.
...
- mac
- linux
- windows
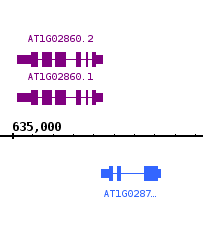
Change the foreground, background, and track label colors
...
to match the following image:
- Validate the color changes
- The main view of IGB is now colored correctly with the three colors you chose.
- mac
- linux
- windows
Change the track label font size to 20.0.
.
...
- The font increased appropriately.
- mac
- linux
- windows
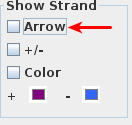
Uncheck the
...
Arrow option in the Show Strand box.
...
- The barbed wire disappeared from the intron sections of gene models
- Select the ".
- mac
- linux
- windows
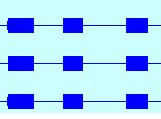
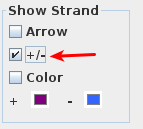
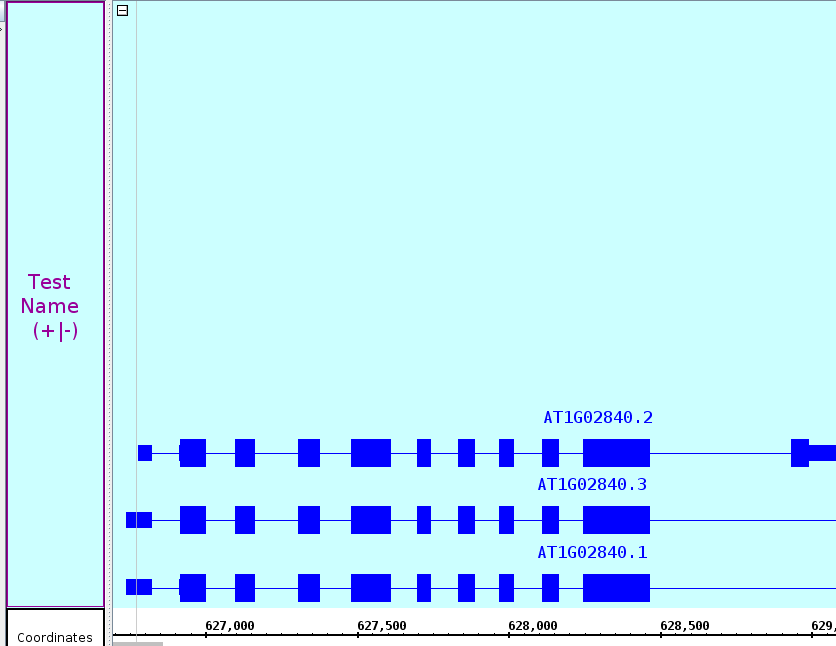
Check the +/-
...
option in the Show Strand box to combine the plus and minus strands.
...
- The two strands are now one.
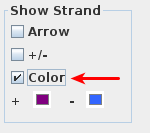
- Select "Color" option under "Show Strand"
...
- mac
- linux
- windows
Check the Color option in the Show Strand box.
- The gene models updated to the colors indicated
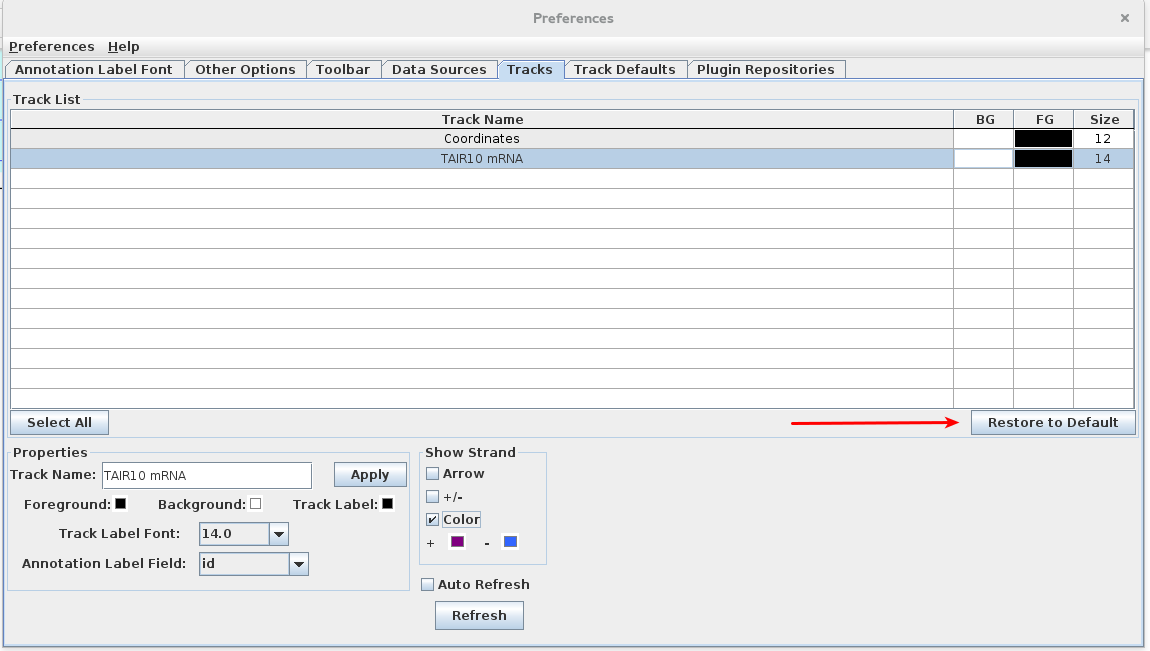
- Select "Restore to Default" option
- Validate that all settings changed in this section (Track Style Preferences) .
- mac
- linux
- windows
Click Restore to Default in the Tracks tab in Preferences.
- All settings changed in the Tracks tab are restored to their original values.
- mac
- linux
- windows
...
Adding and Removing Tracks
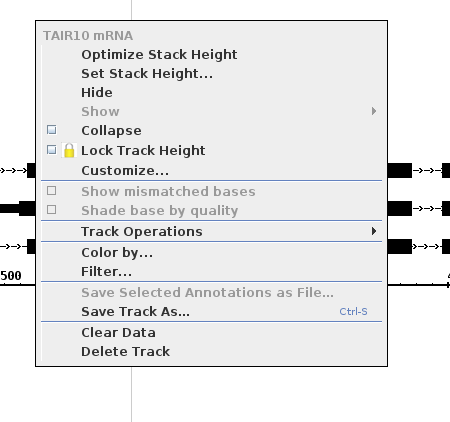
Right click on the white space surrounding the gene models
...
.
- The context menu looks as follows
...
- like the one pictured above.
- mac
- linux
- windows
Select
...
Delete Track
...
.
...
- The track disappeared.
- mac
- linux
- windows
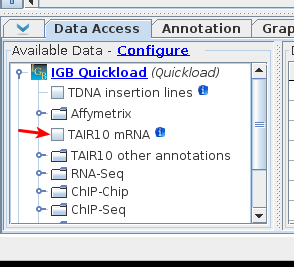
Add the TAIR10 mRNA track back in
...
via the Available Data section.
- The track will reappear reappeared with no data loaded. Select the Load Data button to reload the data.
...
- mac
...
- linux
- windows