General Function Checklist
- All Default DataProviders appear in the DataProviderManagementGui table
...
- Observation: The table lists several Quickload sites. (They may be different from those shown below.) If any are shown with a red background, that means that there is a problem with the site - IGB can't reach it. This is a bug and should be reported.
- Ensure that the edit button in the Data Sources window allows a user to modify an app repository URL.
...
...
Default Data Providers
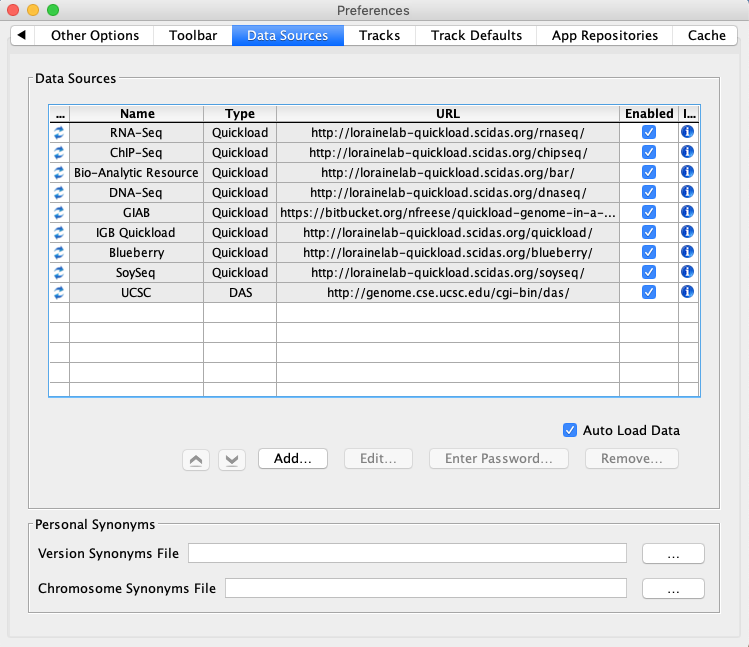
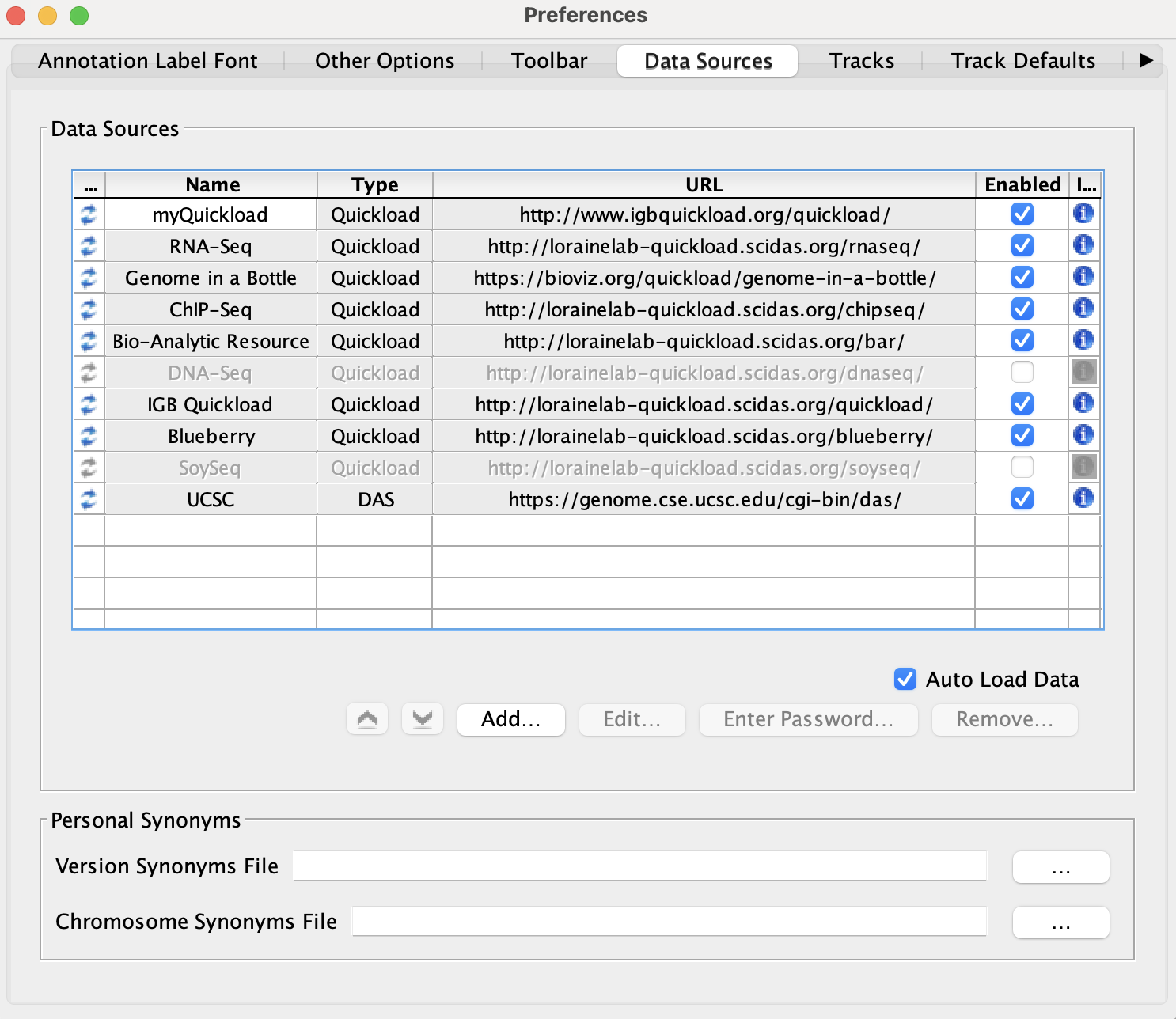
Select File -> Preferences -> Data Sources tab.
- Default data providers (e.g., RNA-Seq, ChIP-Seq, DNA-Seq, etc.) appear in the Data Sources table.
- mac
- linux
- windows
- None of the rows have a red or yellow background.
- mac
- linux
- windows
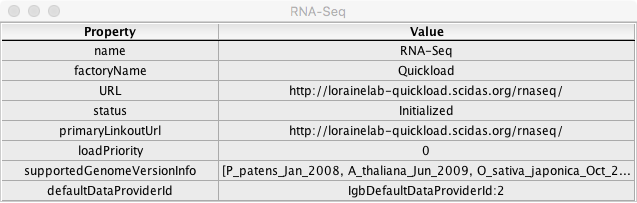
Click on the icon in the Information column for RNA-Seq.
- A window opened with fields and values similar to the example below:
- mac
- linux
- windows
- The data listed in the Value column are specific to the RNA-Seq data source.
- Confirm the following species / genome versions are available Arabidopsis thaliana / A_thaliana_Jun2009Observation: Available Data Tree appears with several sites. (They mac
- linux
- windows
Close this information window, then click on the icon for UCSC.
- The data listed in the Value column updated appropriately and are specific to the UCSC data source.
- mac
- linux
- windows
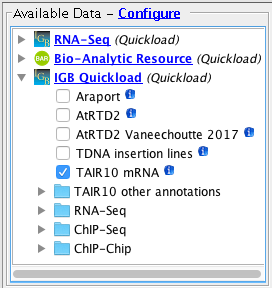
Open the A_thaliana_Jun_2009 genome.
- Several Quickloads are present in the Available Data section (they may be different from those shown below).
...
...
- mac
- linux
- windows
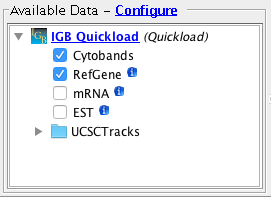
Open the H_sapiens_Dec_
...
2013 genome.
- Several Quickloads are present in the Available Data section (they may be different from those shown below).
...
- mac
- linux
- windows
- Important:
...
- The Cytobands track is visible
...
- Load a bed file, a bam file, a bedgraph file, and some sequence data to ensure basic file parsing from the data providers is working as expected. Important: Load these by selecting the checkboxes in the Data Access folder hierarchy, not from URLs displayed in a Web browser. To get examples of each file type, go the the Arabidopsis thaliana June 2009 genome. Under RNA-Seq there are several folders labeled by experiment code and with a human-friendly name. Within those folders, there are folders labeled Reads, Graph, and Junctions. The Reads folder has bam files. The Graph folder has bedgraph files. The Junctions folder has bed files. Select one file from each folder, zoom in to show only several thousand bases, and click Load Data button at the top right of the IGB window. Click the Load Sequence button to load sequence and zoom in to make sure you can see individual letter (A, C, T, G) under the coordinate axis.
- Bed File
- Bam File
- Bedgraph File
- Load Sequence
- Load a few tracks from the UCSC DAS data source. Note: The UCSC DAS data source is not activated by default - you need to activate it in the Data Sources tab of the Preference window. Once this data source is active, it should be visible on Human/Mouse species.
...
- mac
- linux
- windows
- Navigate to chr1:45,691,287-45,691,329
- Click Load Sequence.
- Individual base pairs (ACGT) are visible and colored consistently along the coordinates track.
- mac
- linux
- windows
Adding/Removing/Editing Data Providers
- Select File -> Preferences -> Data Sources tab.
- Click Add...
- Name: myQuickload
- Type: Quickload
- URL: http://www.igbquickload.org/
...
- The quickload site shows up in the "Available Data" tree
- The data sets listed in the tree can be loaded
- Confirm restarting IGB does not cause the newly added site to be forgotten
...
- Click Submit.
- Close Preferences.
- myQuickload is present in the Available Data section
- mac
- linux
- windows
- Close and re-open IGB.
- Select File -> Preferences -> Data Sources tab.
- myQuickload is still present in the Data Sources table.
- mac
- linux
- windows
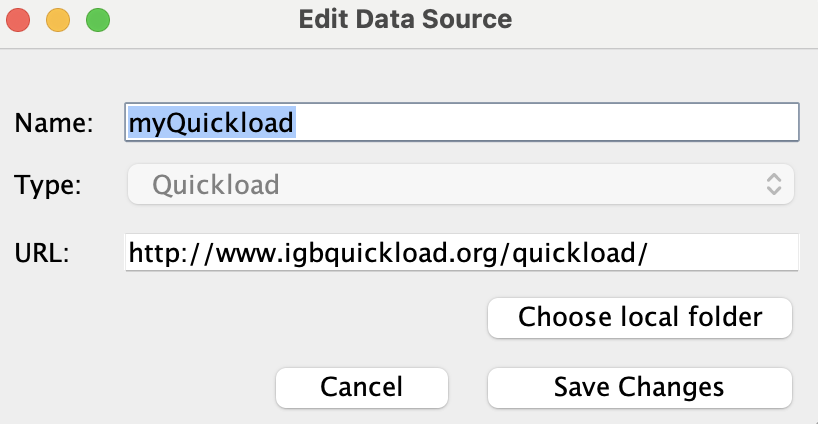
Select myQuickload and click Edit...
- The Edit Data Source window opened.
- mac
- linux
- windows
Click Choose local folder.
- The user's file chooser opens correctly.
- mac
- linux
- windows
Change the Name of myQuickload to myQuickloadv2, then click Save Changes.
- The name of myQuickload correctly updated to myQuickloadv2 in both the Data Sources table and the Available Data section.
- mac
- linux
- windows
- Select myQuickloadv2.
- Click Remove...
- The myQuickloadv2 data source was removed from both the Data Sources table and the Available Data section.
- mac
- linux
- windows
Secured Data Providers
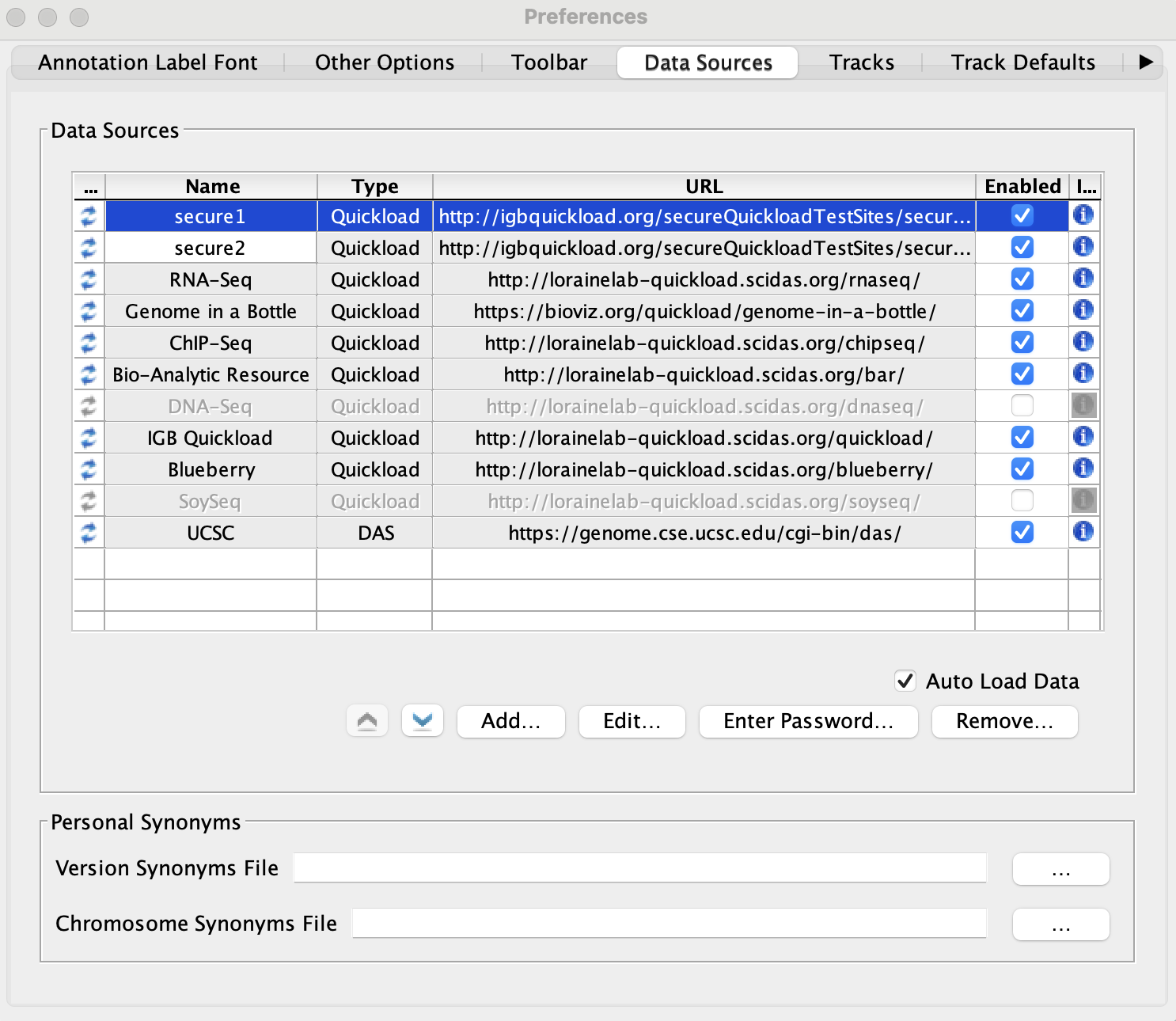
Click Add... and add the following two Quickload sites:
- Name: secure1
- Type: Quickload
- URL: http://igbquickload.org/secureQuickloadTestSites/secureSiteTest
...
- guest/guest
- Click Submit. A prompt should appear asking for a username and password:
- Username: guest
- Password: guest
- Make sure Save Password is checked, then click OK.
- Click Submit. A prompt should appear asking for a username and password:
- Name: secure2
- Type: Quickload
- URL: http://igbquickload.org/secureQuickloadTestSites/secureSiteTest2
...
- guest2/guest2
...
- Click Submit. A prompt should appear asking for a username and password:
- Username: guest2
- Password: guest2
- Make sure Save Password is checked, then click OK.
- Click Submit. A prompt should appear asking for a username and password:
- The two secured Quickloads are added to the Data Sources table and are not highlighted yellow or red.
- mac
- linux
- windows
Close Preferences and open the A_thaliana_Jun_2009 genome
...
.
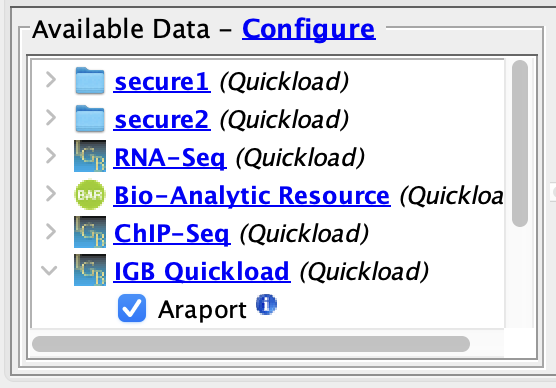
- Confirm the The two sites secured Quickloads are listed in the "Available Data" tree and data can be loaded from each siteConfirm you Available Data section.
- mac
- linux
- windows
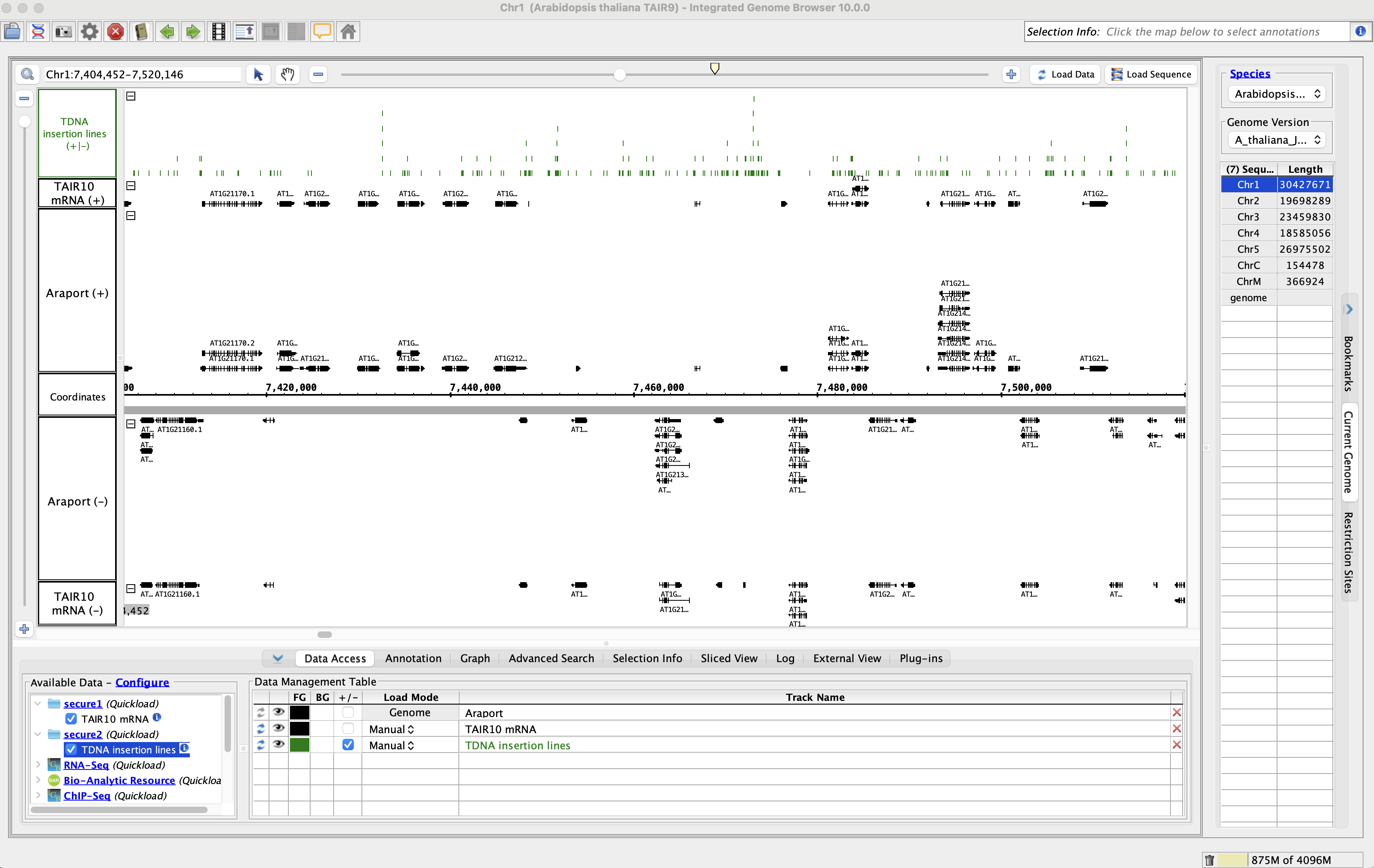
- Add the TAIR10 mRNA data from the secure1 Quickload.
- Add the TDNA insertion lines data from the secure2 Quickload.
- Go to Chr1:7,404,452-7,520,146
- Click Load Data.
- Data loaded from both of the secured Quickloads.
- mac
- linux
- windows
- You were not prompted for a password for any site more than a single time per session
- Confirm restarting IGB does not cause the newly added sites to be forgotten
- mac
- linux
- windows
Restart IGB and open the A_thaliana_Jun_2009 genome.
- The two secured Quickloads are still listed in the Available Data section.
- mac
- linux
- windows
- You were not prompted for your password again on for either of the secured Quickloads upon restarting IGB.
...
- mac
- linux
- windows