General Function Checklist
Default Data Providers
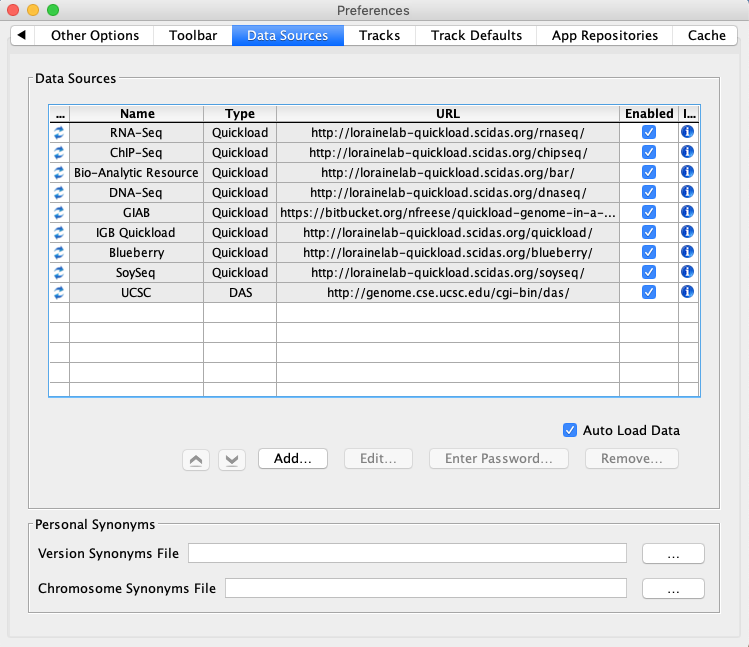
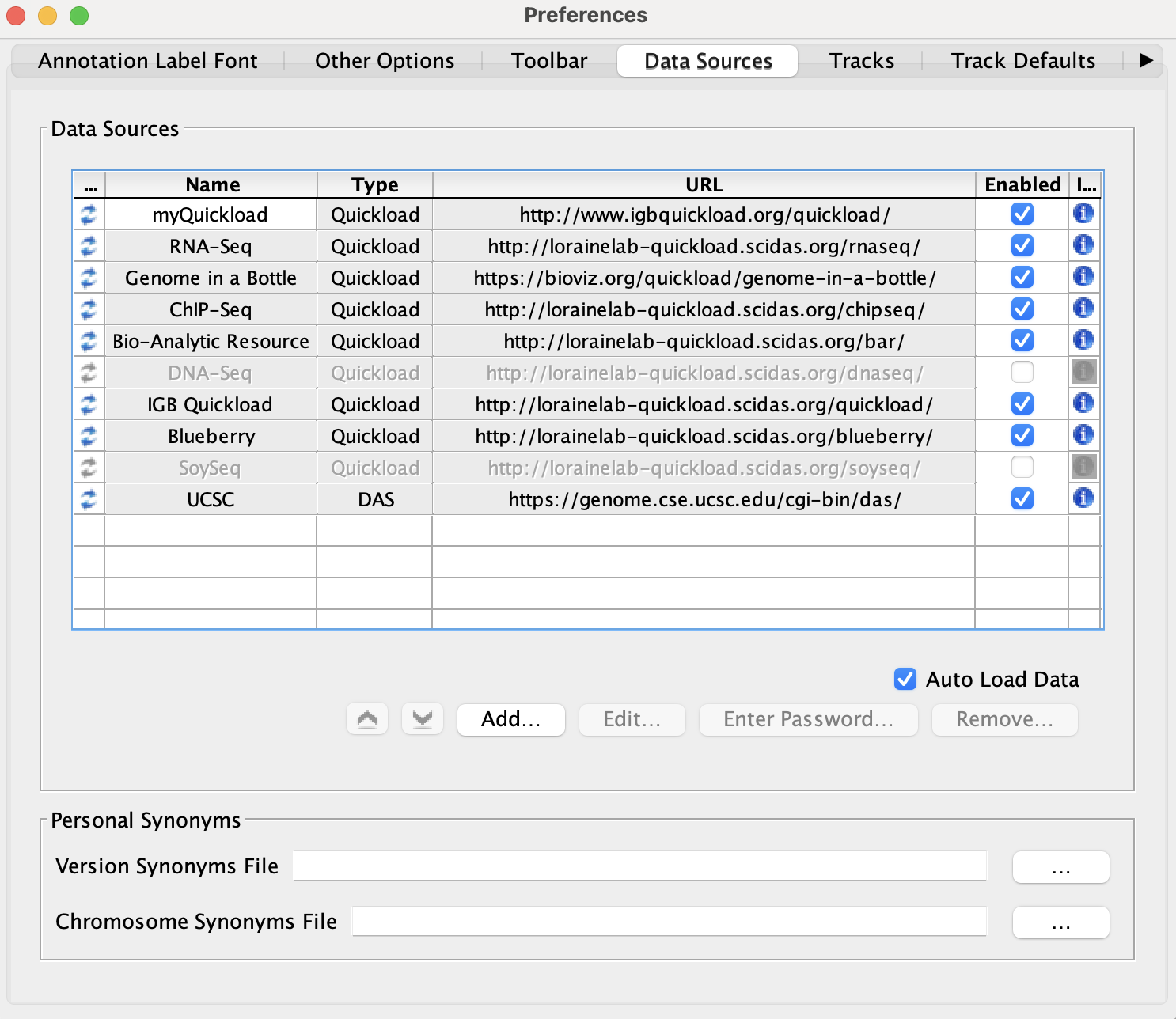
Select File -> Preferences -> Data Sources tab.
- Default data providers (e.g., RNA-Seq, ChIP-Seq, DNA-Seq, etc.) appear in the Data Sources table.
- mac
- linux
- windows
- None of the rows have a red or yellow background.
- mac
- linux
- windows
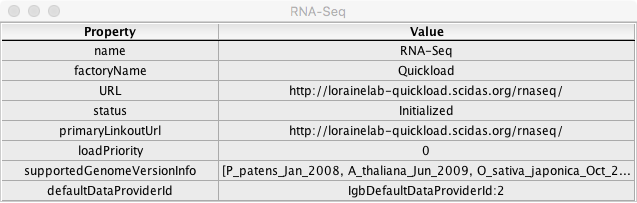
Click on the icon in the Information column for RNA-Seq.
- A window opened with fields and values similar to the example below:
- mac
- linux
- windows
- The data listed in the Value column are specific to the RNA-Seq data source.
- mac
- linux
- windows
Close this information window, then click on the icon for Genome in a Bottle.
- The data listed in the Value column updated appropriately and are specific to the Genome in a Bottle data source.
- mac
- linux
- windows
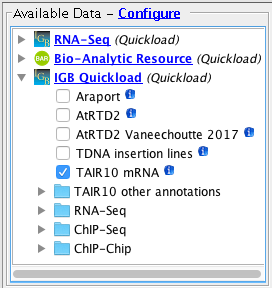
Open the A_thaliana_Jun_2009 genome.
- Several Quickloads are present in the Available Data section (they may be different from those shown below).
- mac
- linux
- windows
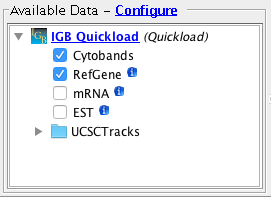
Open the H_sapiens_Dec_2013 genome.
- Several Quickloads are present in the Available Data section (they may be different from those shown below).
- mac
- linux
- windows
- Important: The Cytobands track is visible
- mac
- linux
- windows
- Navigate to chr1:45,691,287-45,691,329
- Click Load Sequence.
- Individual base pairs (ACGT) are visible and colored consistently along the coordinates track.
- mac
- linux
- windows
Adding/Removing/Editing Data Providers
- Select File -> Preferences -> Data Sources tab.
- Click Add...
- Name: myQuickload
- Type: Quickload
- URL: http://www.igbquickload.org/quickload/
- Click Submit.
- Close Preferences.
- myQuickload is present in the Available Data section
- mac
- linux
- windows
- Close and re-open IGB.
- Select File -> Preferences -> Data Sources tab.
- myQuickload is still present in the Data Sources table.
- mac
- linux
- windows
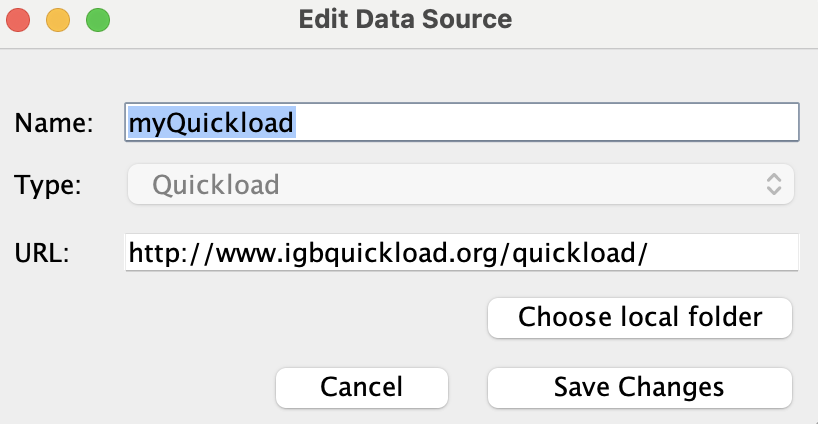
Select myQuickload and click Edit...
- The Edit Data Source window opened.
- mac
- linux
- windows
Click Choose local folder.
- The user's file chooser opens correctly.
- mac
- linux
- windows
Change the Name of myQuickload to myQuickloadv2, then click Save Changes.
- The name of myQuickload correctly updated to myQuickloadv2 in both the Data Sources table and the Available Data section.
- mac
- linux
- windows
- Select myQuickloadv2.
- Click Remove...
- The myQuickloadv2 data source was removed from both the Data Sources table and the Available Data section.
- mac
- linux
- windows
Secured Data Providers
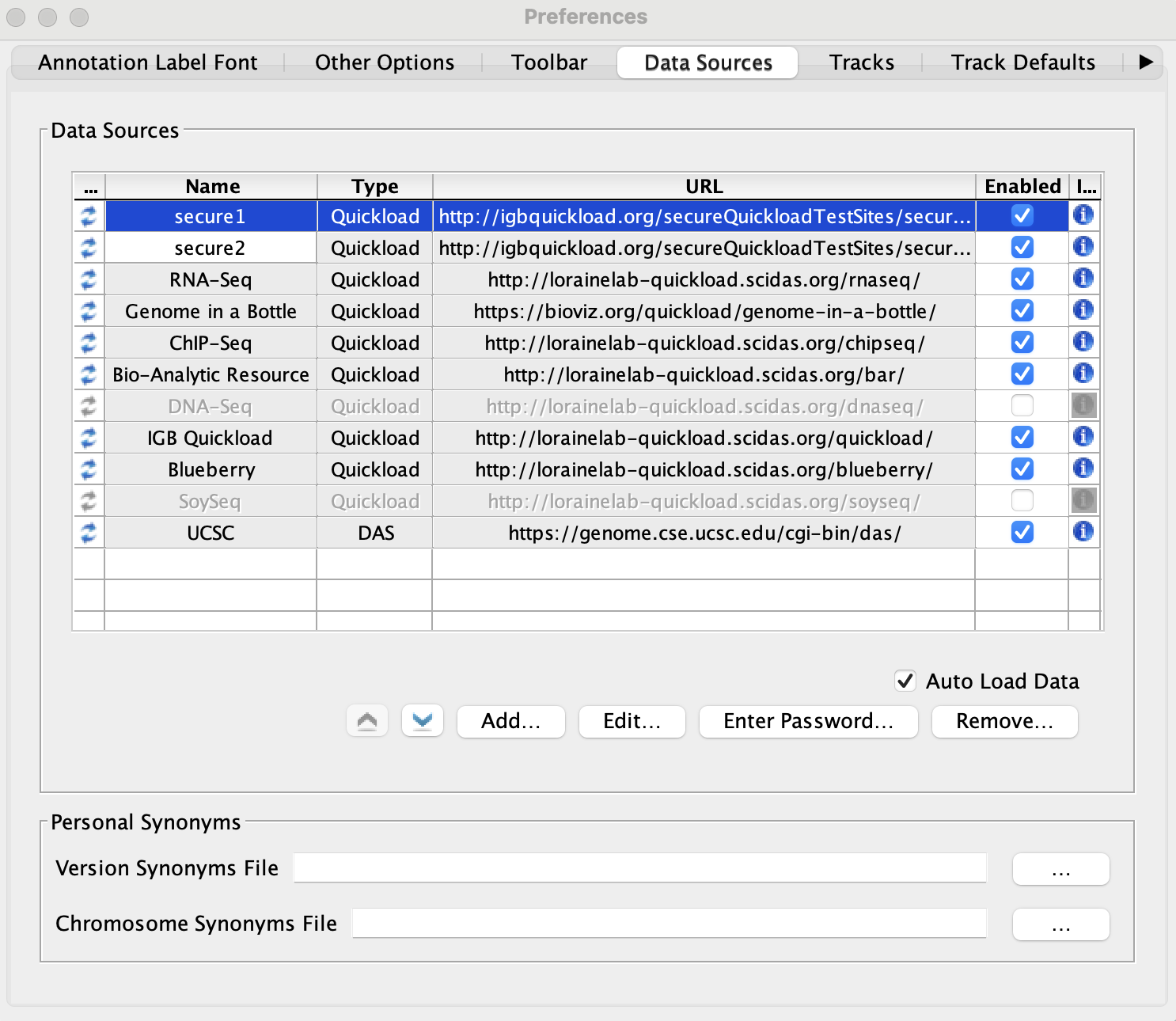
Click Add... and add the following two Quickload sites:
- Name: secure1
- Type: Quickload
- URL: http://igbquickload.org/secureQuickloadTestSites/secureSiteTest
- Click Submit. A prompt should appear asking for a username and password:
- Username: guest
- Password: guest
- Make sure Save Password is checked, then click OK.
- Click Submit. A prompt should appear asking for a username and password:
- Name: secure2
- Type: Quickload
- URL: http://igbquickload.org/secureQuickloadTestSites/secureSiteTest2
- Click Submit. A prompt should appear asking for a username and password:
- Username: guest2
- Password: guest2
- Make sure Save Password is checked, then click OK.
- Click Submit. A prompt should appear asking for a username and password:
- The two secured Quickloads are added to the Data Sources table and are not highlighted yellow or red.
- mac
- linux
- windows
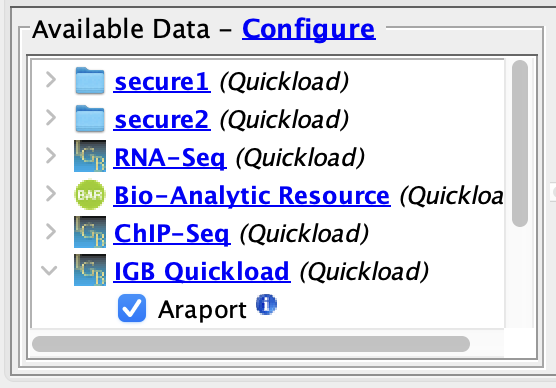
Close Preferences and open the A_thaliana_Jun_2009 genome.
- The two secured Quickloads are listed in the Available Data section.
- mac
- linux
- windows
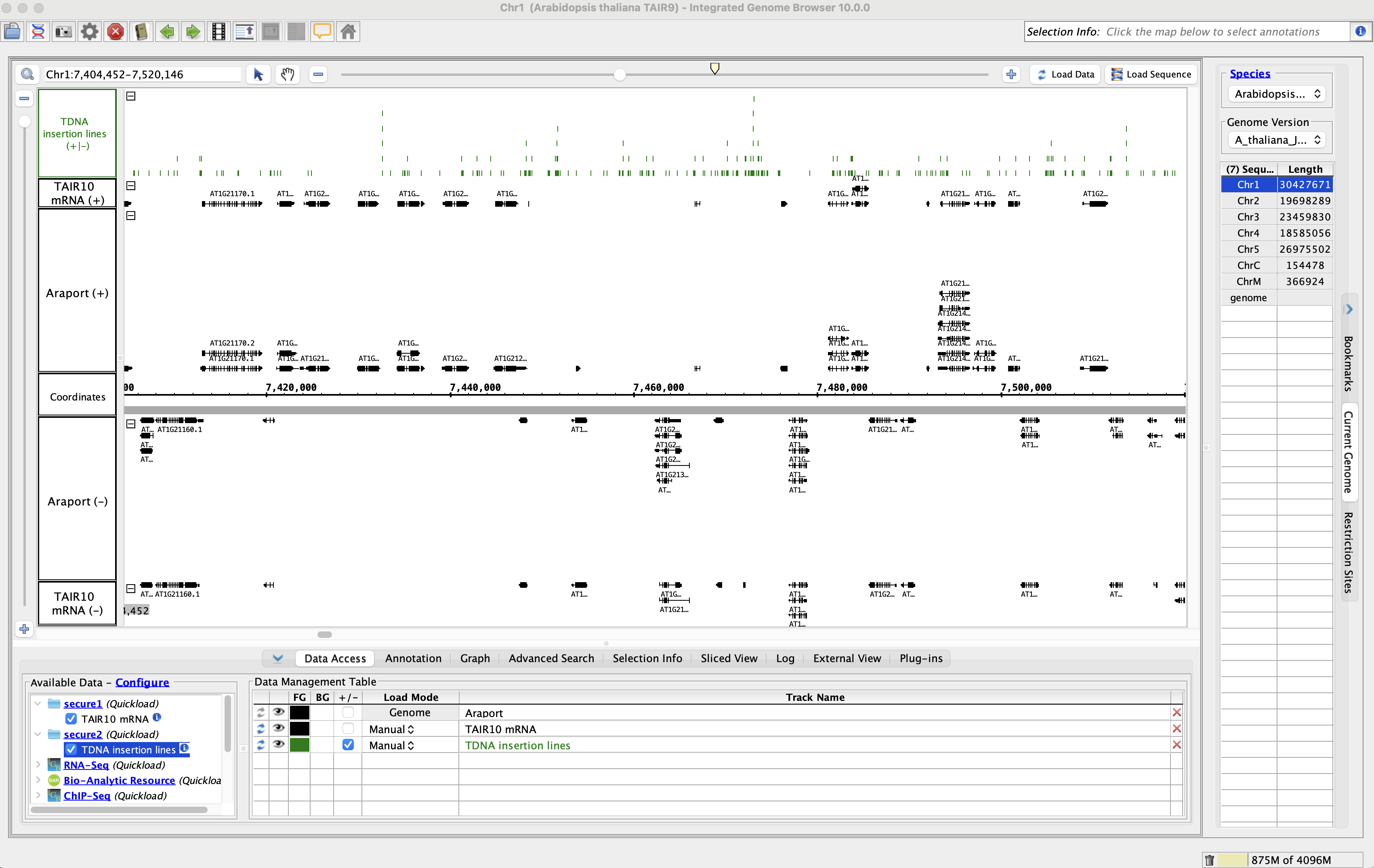
- Add the TAIR10 mRNA data from the secure1 Quickload.
- Add the TDNA insertion lines data from the secure2 Quickload.
- Go to Chr1:7,404,452-7,520,146
- Click Load Data.
- Data loaded from both of the secured Quickloads.
- mac
- linux
- windows
- You were not prompted for a password for either of the secured Quickloads.
- mac
- linux
- windows
Restart IGB and open the A_thaliana_Jun_2009 genome.
- The two secured Quickloads are still listed in the Available Data section.
- mac
- linux
- windows
- You were not prompted for your password again for either of the secured Quickloads upon restarting IGB.
- mac
- linux
- windows