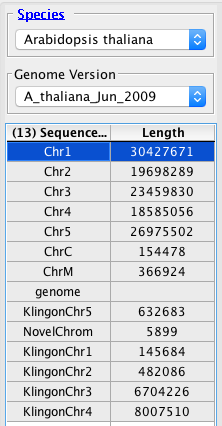
General Function Checklist
See this page in the users guide: Personal Synonyms
Chromosome synonyms
Here is an example chromosome synonyms file: chromosome.txt
KlingonChr1 Chr1
KlingonChr2 Chr2
KlingonChr3 Chr3
KlingonChr4 Chr4
KlingonChr5 Chr5
Here is the head of an example bed file, made by taking segments from the Athe A_thaliana_Jun_2009 gene annotations and replacing the chromosome names: KlingonGenes.bed
NovelChrom 3630 5899 AT1G01010.1 0 + 3759 5630 0 6 283,281,120,390,153,461, 0,365,855,1075,1543,1808,
KlingonChr1 5927 8737 AT1G01020.1 0 - 6914 8666 0 10 336,633,76,67,86,74,46,90,48,167, 0,509,1229,1456,1636,1834,2014,2308,2489,2643,
KlingonChr1 6789 8737 AT1G01020.2 0 - 7314 8666 0 8 280,294,86,74,46,90,48,167, 0,367,774,972,1152,1446,1627,1781,
KlingonChr1 11648 13714 AT1G01030.1 0 - 11863 12940 0 2 1525,380, 0,1686,
KlingonChr1 23145 31227 AT1G01040.1 0 + 23518 31079 0 20
etc.
Notice that there is one entry for a chromosome (NovelChrom) that is not in the synonyms file.As per the directions in Personal Synonyms, add this chromosome.txt file as a chromosome synonyms file.
- Navigate to the Data Sources tab in Preferences.
- Click ... next to the Chromosome Synonyms File field.
- Add the chromosome.txt file.
- Restart IGB.
- Open the A_thaliana_Jun_2009 genome.
- Load the KlingonGenes.bed file from the Available Data section.
- Mac
- Windows
- Linux
- Ensure the Load Mode in the Data Access tab at the bottom of IGB is set to Genome.
Good.
Bad.
- The annotations from KlingonGenes.bed appear on the same chromosomes as the automatically loaded annotations.
- (If IGB creates new chromosomes labeled "KlingonChr1" etc, then the link between chromosome "KlingonChr5" and "Chr5" is not functioning.)
- mac
- linux
- windows
Genome version synonyms
Review the user guide page: Use synonyms.txt to link genome version names to each other.
How to make the test file
(These instructions are a work in progress
.)
You can download the test file I made: myQL
Unzip that folder, and continue to How to use the test file.
The following instructions here should be viewed as a "fall back".
- Make an empty folder called "myQL".
- In the myQL folder, create a plain text file called contents.txt file with this line (it is a tab between 2018 and My; do not include the bullet):
- A_new_Jun_2018 My new genome as a test
- In myQL, create a folder called "A_new_Jun_2018"
- In A_new_Jun_2018, create an annots.xml file with this text:
<files>
<file name="KlingonGenes.bed" title="new/KlingonGenes" />
</files>
- In the A_new_Jun_2018 folder, save the KlingonGenes.bed file (see Chromosome synonyms).
- In the myQL folder, create a folder called "synsFileNotInUse"
- In the synsFileNotInUse folder, create a text file called synonyms.txt with this text (this is two terms, separated by a tab):
- A_thaliana_Jun_2009 A_new_Jun_2018
The synonyms file will not "work" from this location. This is just its starting position in the test process.
How to use the test file
- Any platform
- Download the myQL test file here: myQL
- Reset preferences to give IGB a clean slate.
- From the IGB home screen, click the species Species pull down. Verify there
- There is NO genome for
- A_new
- in the Species pull down.
- mac
- linux
- windows
- In the Data Sources tab in Preferences, add myQL as a quickload Quickload site.
- From the IGB home screen, click the species Species pull down. Verify there
- There IS a genome for
- A_new
- in the Species pull down.
- mac
- linux
- windows
Open A_new_Jun_2018.
- The myQL Quickload appears in the Available Data section.
- mac
- linux
- windows
- Add KlingonGenes from the myQL Quickload.
- Click Load Data.
- The KlingonGenes data loaded and Klingon chromosomes are now listed in the panel at the right
- Verify that there is no myQL folder in the data access
- IGB should prompt you to restart. Restart IGB.
- Verify that there is NO "A_new" genome.
- In the Arabidopsis genome, under data access, verify that myQL is available.
- .
- mac
- linux
- windows
Open A_thaliana_Jun_2009.
- There is NO myQL Quickload in the Available Data section.
- mac
- linux
- windows
In the Data Sources tab in Preferences, add synonyms.txt file (found in myQL > synsFileNotInUse) as a version synonyms file.
- IGB prompted you to restart.
- mac
- linux
- windows
Restart IGB.
- There is NO genome for A_new in the Species pull down.
- mac
- linux
- windows
Open A_thaliana_Jun_2009.
- The myQL Quickload appears in the Available Data section.
- mac
- linux
- windows
- Add KlingonGenes from the myQL Quickload.
- Click Load Data.
- The KlingonGenes data loaded and Klingon chromosomes are now listed in the panel at the right.
- mac
- linux
- windows
- Reset preferences to default.
- Move In your File Chooser, move the synonyms.txt file from the synsFileNotInUse folder to the myQL folder.
- Add the myQL quickload Restart IGB.
- In the Data Sources tab in Preferences, add myQL as a Quickload site.Verify that
- There is NO
- genome for A_new
- The myQL folder an the KlingonGenes file are available in the data access panel in the Arabidopsis genome.
- in the Species pull down.
- mac
- linux
- windows
Open A_thaliana_Jun_2009.
- The myQL Quickload appears in the Available Data section.
- mac
- linux
- windows
- Add KlingonGenes from the myQL Quickload.
- Click Load Data.
- The myQL Quickload and the KlingonGenes file were loaded without issue with the A_thaliana_Jun_2009 genome.
- mac
- linux
- windows