| Table of Contents |
|---|
Introduction
Use this tab to view the currently visible region in the Ensembl or UCSC Genome Browsers.
Use this tab to
- See an alternative view on the same data
- Find tracks to download and import into the IGB environment
Both Ensembl and UCSC Genome Bioinformatics sites are accessible via DAS1 data sources in IGB. And the UCSC Genome site has a Table Browser that allows you to download Genome Browser data tracks in formats IGB supports.
So if you see something worth further investigation, you can import the data directly into IGB.
View a region in an external genome browser
Depending on the genome, you can use External View tab to view the currently visible region in the UCSC or Ensembl genome browsers.
...
If you navigate to a new region, click the Update button.
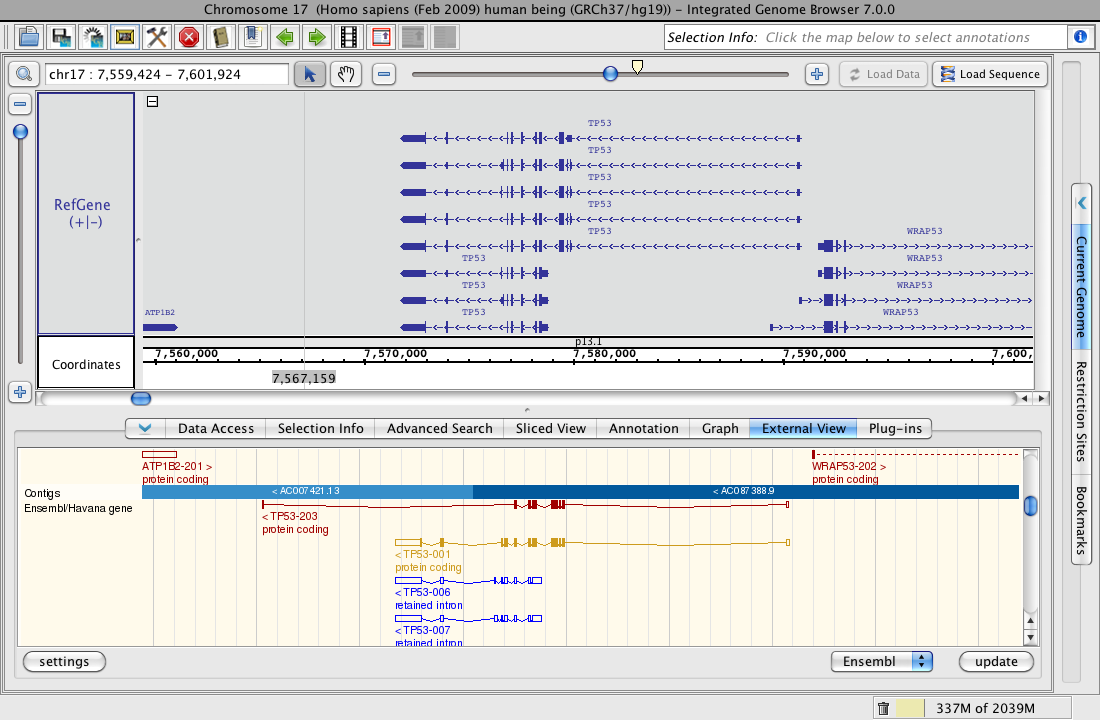
Although the view is not interactive, you can get interesting details about a gene/region, including cross species comparisons, mapped ESTs, SNPs, and/or repeated sequence data, when available.
External View (click to enlarge)
External View showing TP53 in IGB and the Ensembl Genome Browser
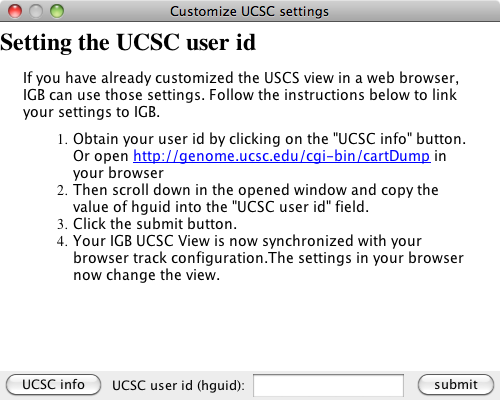
Import UCSC genoe browser settings
If you have used a web browser to access the UCSC server and have changed the UCSC view through the browser, you can import those settings into IGB, permitting IGB to display the view in the same way as the web browser. Click the settings button (blue box in previous image) and follow the directions in the window.
Settings window
Advice on region sizes
While you can access information on very large regions (spanning 10-15 genes), a smaller region or specific gene may return more data due to more details being available. To use ENSEMBL you must have a fairly zoomed in picture (<100,000 bp) and it must be a species that ENSEMBL curates. The same is true for UCSC, although it can handle slightly larger regions of the chromosome. A smaller region or specific gene may return more data due to more details being available.
...