Introduction
Use this tab to view the currently visible region in the UCSC Genome Browser.
UCSC Genome Browser data tracks are also accessible in the Available Data window of the Data Access tab in IGB.
View a region in an external genome browser
Depending on the genome, you can use External View tab to view the currently visible region in an external browser.
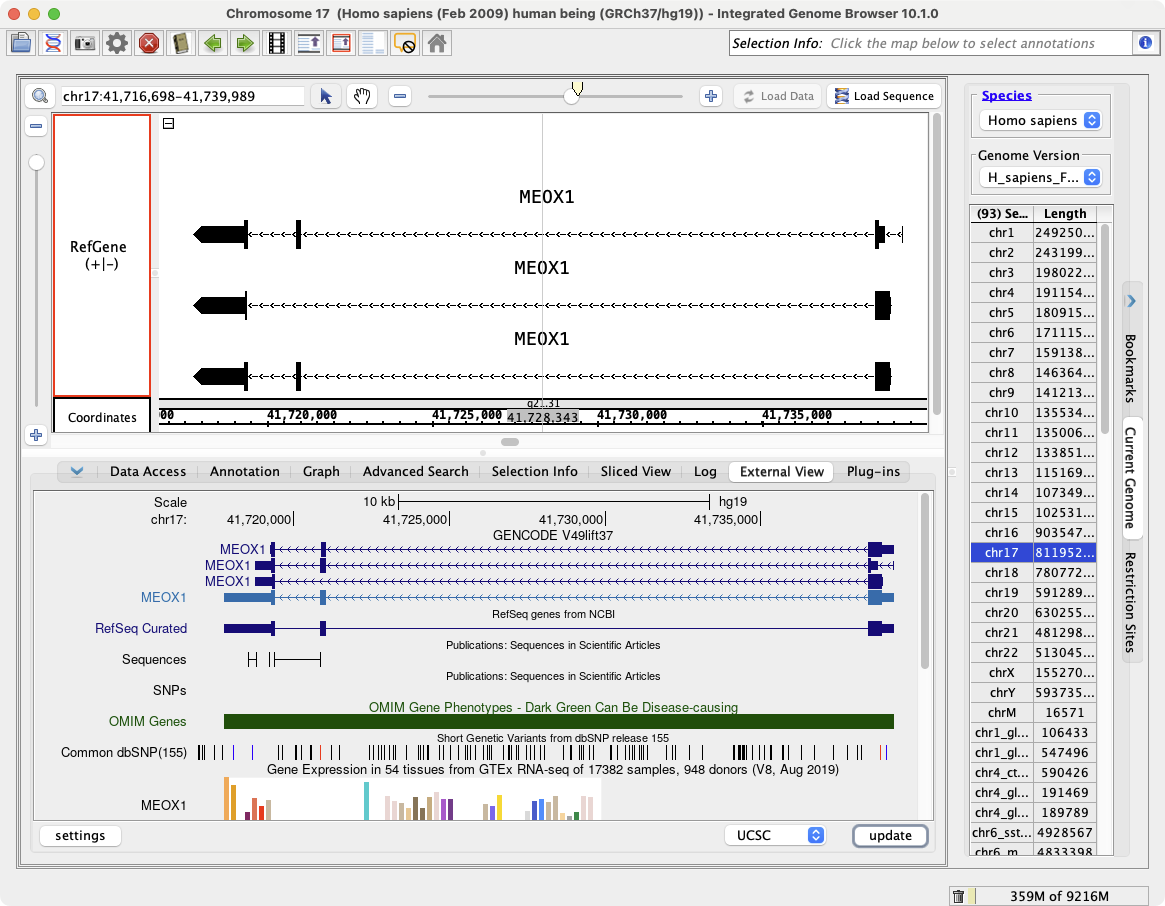
External View showing MEOX1 in IGB and the UCSC Genome Browser
To view a region in the UCSC genome browser
- Select the External View tab
- Click Update
- You will need to click the Update button each time you navigate to a new region
Set a UCSC user id
The first time you use the External View with UCSC you will need to set the UCSC user id (hguid).
1) Select the External View tab
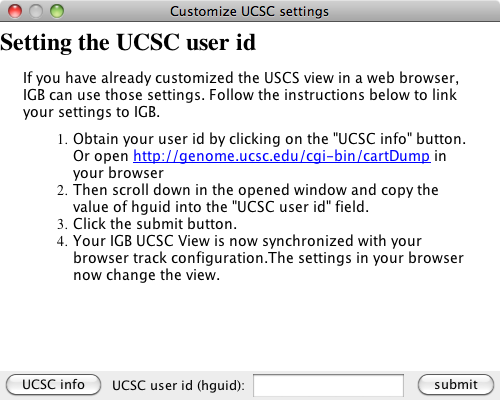
2) Click on the settings button, a new window should open
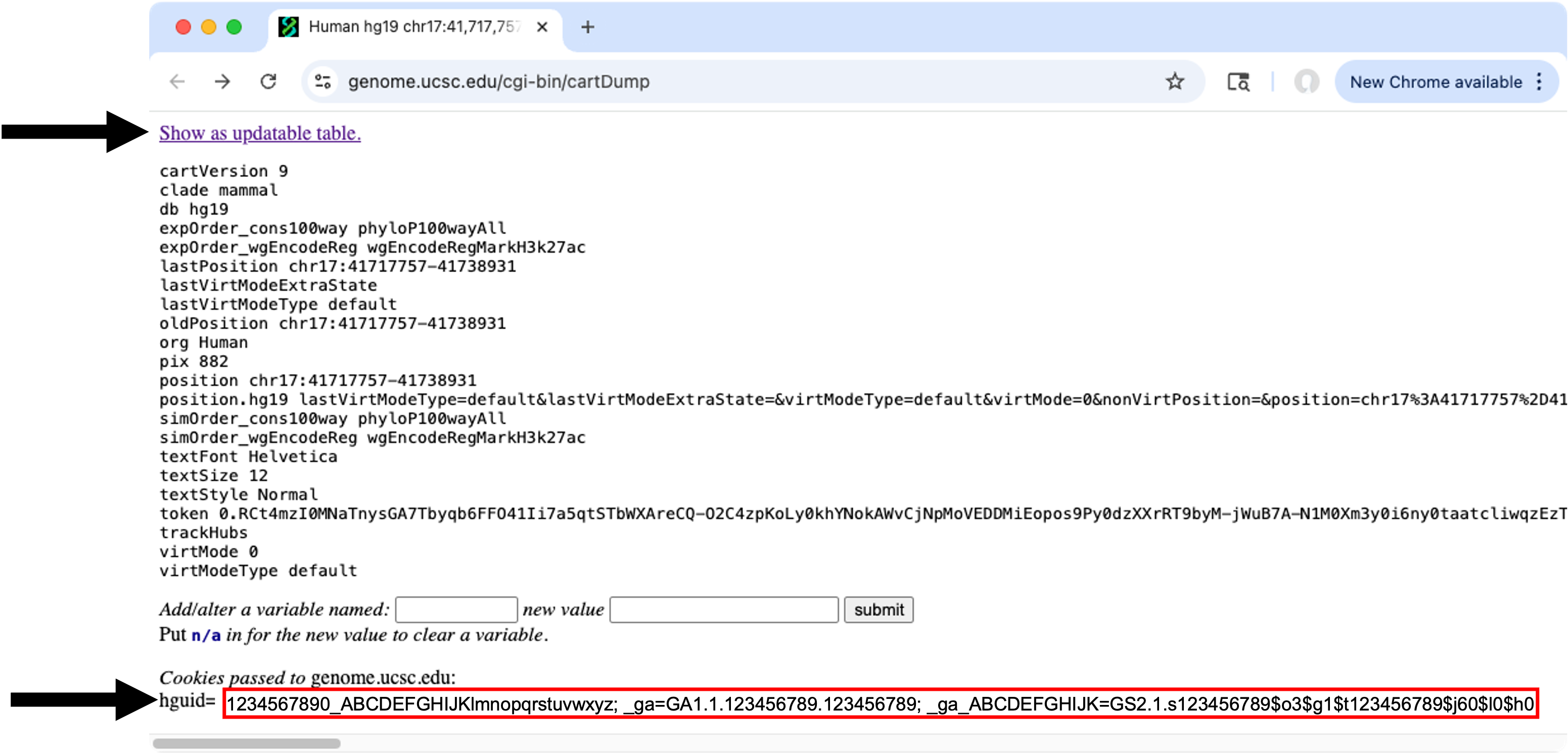
3) In the Customize UCSC settings window, click on the UCSC info button, a web browser window should open
4) Click Show as updatable table.
5) In the browser window, copy the value after hguid=
6) Paste the value into the UCSC user id (hguid) textbox in the Customize UCSC settings window
7) Click the submit button
Import UCSC genome browser settings
If you have used a web browser to access the UCSC server and have changed the UCSC view through the browser, you can import those settings into IGB, permitting IGB to display the view in the same way as the web browser. Click the settings button (blue box in previous image) and follow the directions in the window.
Settings window
Advice on region sizes
While you can access information on very large regions (spanning 10-15 genes), a smaller region or specific gene may return more data due to more details being available. To use ENSEMBL you must have a fairly zoomed in picture (<100,000 bp) and it must be a species that ENSEMBL curates. The same is true for UCSC, although it can handle slightly larger regions of the chromosome. A smaller region or specific gene may return more data due to more details being available.
Larger regions take longer to load.