...
Soft-clipping of reads is done during alignment to mask the beginning or end of a read that does portions of the reads that do not align to the reference sequence. Soft
Visualizing soft-clipping of reads can occur due to un-trimmed can be useful for identifying contaminating adapter sequences or detecting structural variationvariants.
How to view soft-clipping in IGB
Note: Soft-clipped sections of reads are not visible in versions of IGB prior to 9.1
To view your own data with soft-clipping within IGB, first load a BAM filein IGB:
- Open Add your BAM file to IGB.
- Zoom in on the gene or region of interest.
- Click Load Data.
To view the example soft-clipping data in the images below:
- Open the human genome (H_sapiens_Dec_2013) in IGB.
- Navigate to: chr1:3,320-26,768
- Hide the RefSeq Curated annotation track by right-clicking and selecting Hide.
- Download the two files below:
- In IGB, select File > Open File...
- Select the pacBio.bam file and click Open.
- Click Load Sequence to load sequence data.
- Click Load Data to load data into it.
Soft-clipping is enabled by default in IGB, with the soft-clipped section of the read displayed in gray.
...
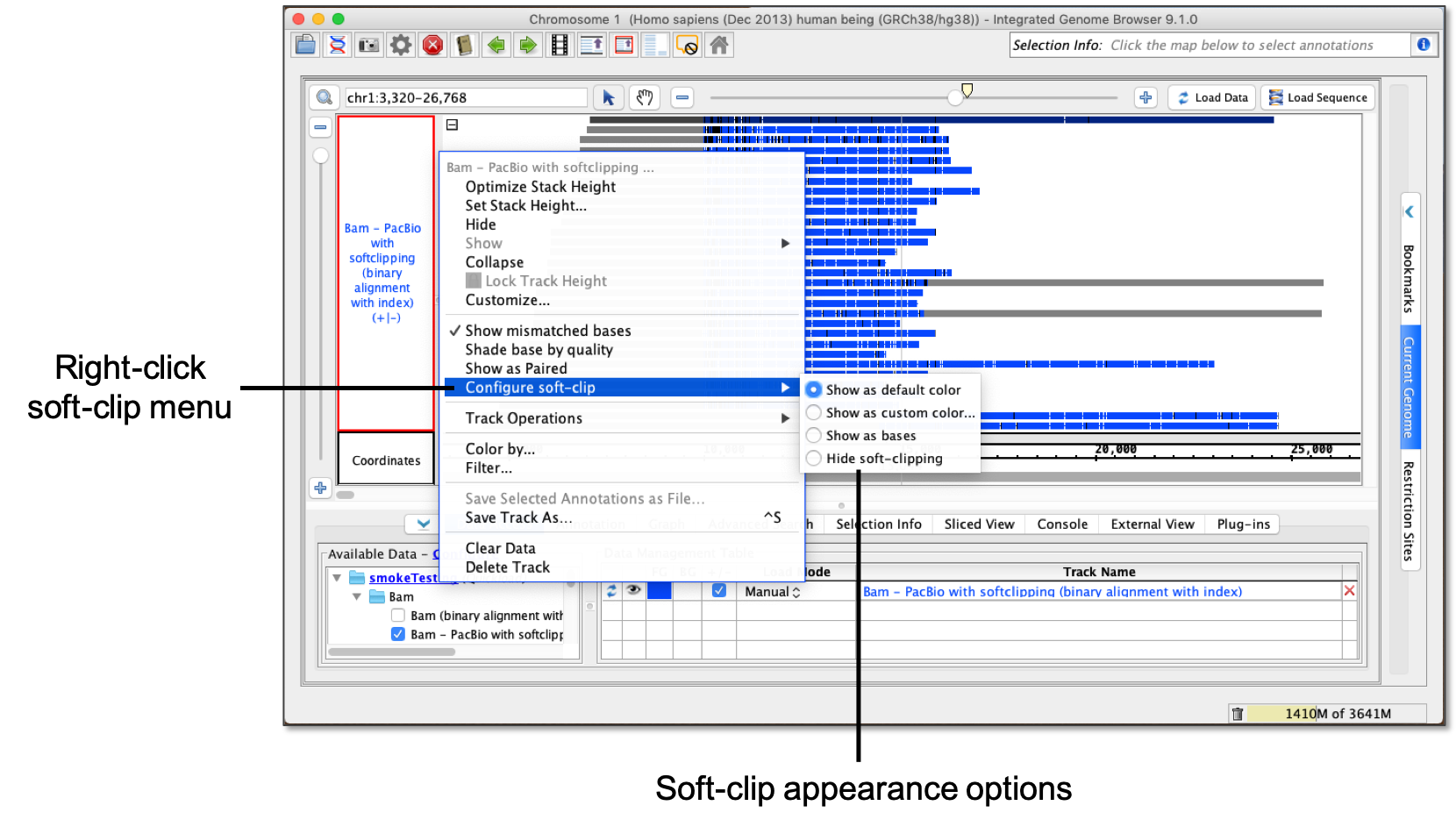
- Right-click the reads track label.
- Select Configure soft-clip.
Options for viewing soft-clipping include:
- Show as default color - softSoft-clipping shows as a default gray.
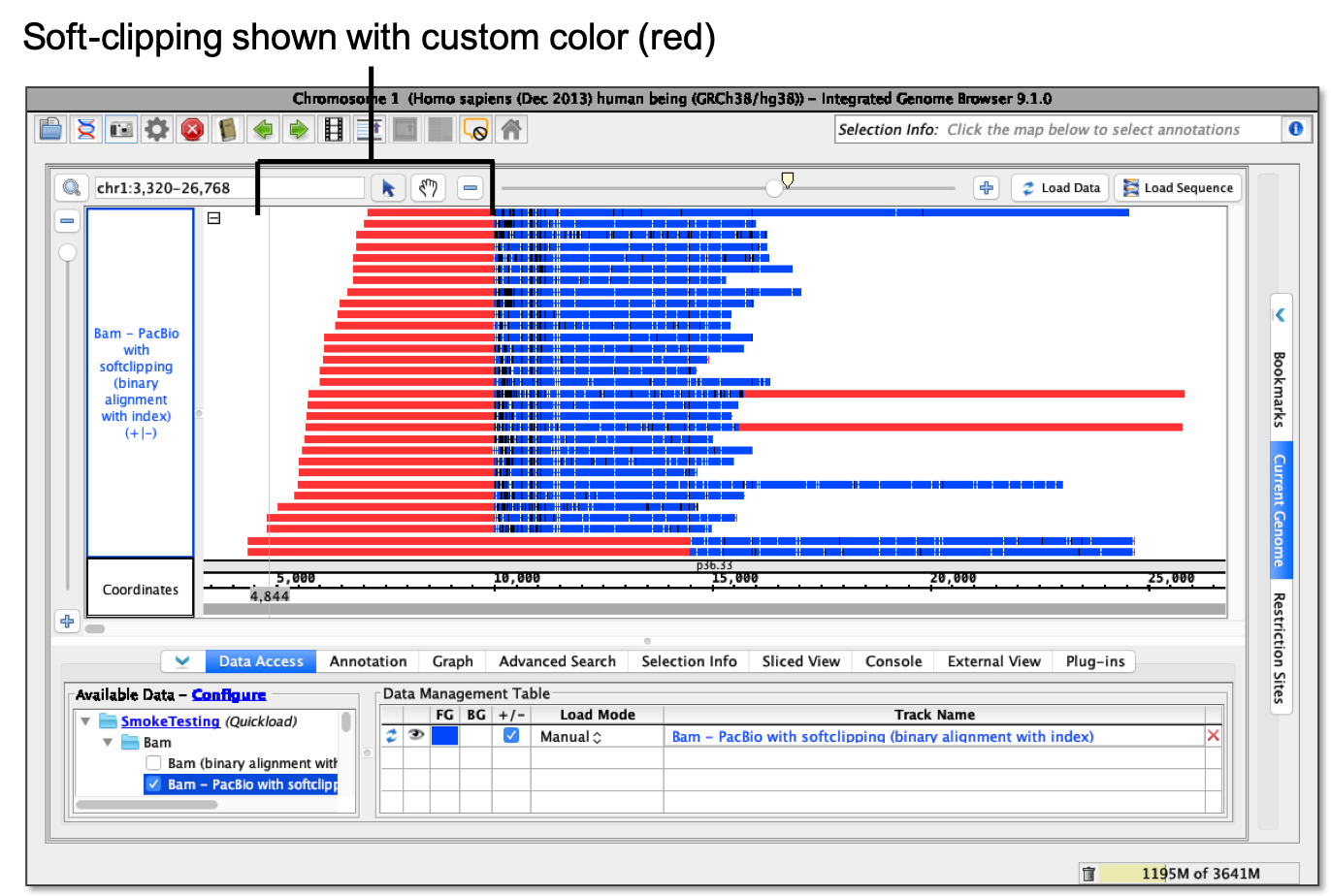
- Show as custom color... - using Using a color picker, soft-clipping shows as selected color.
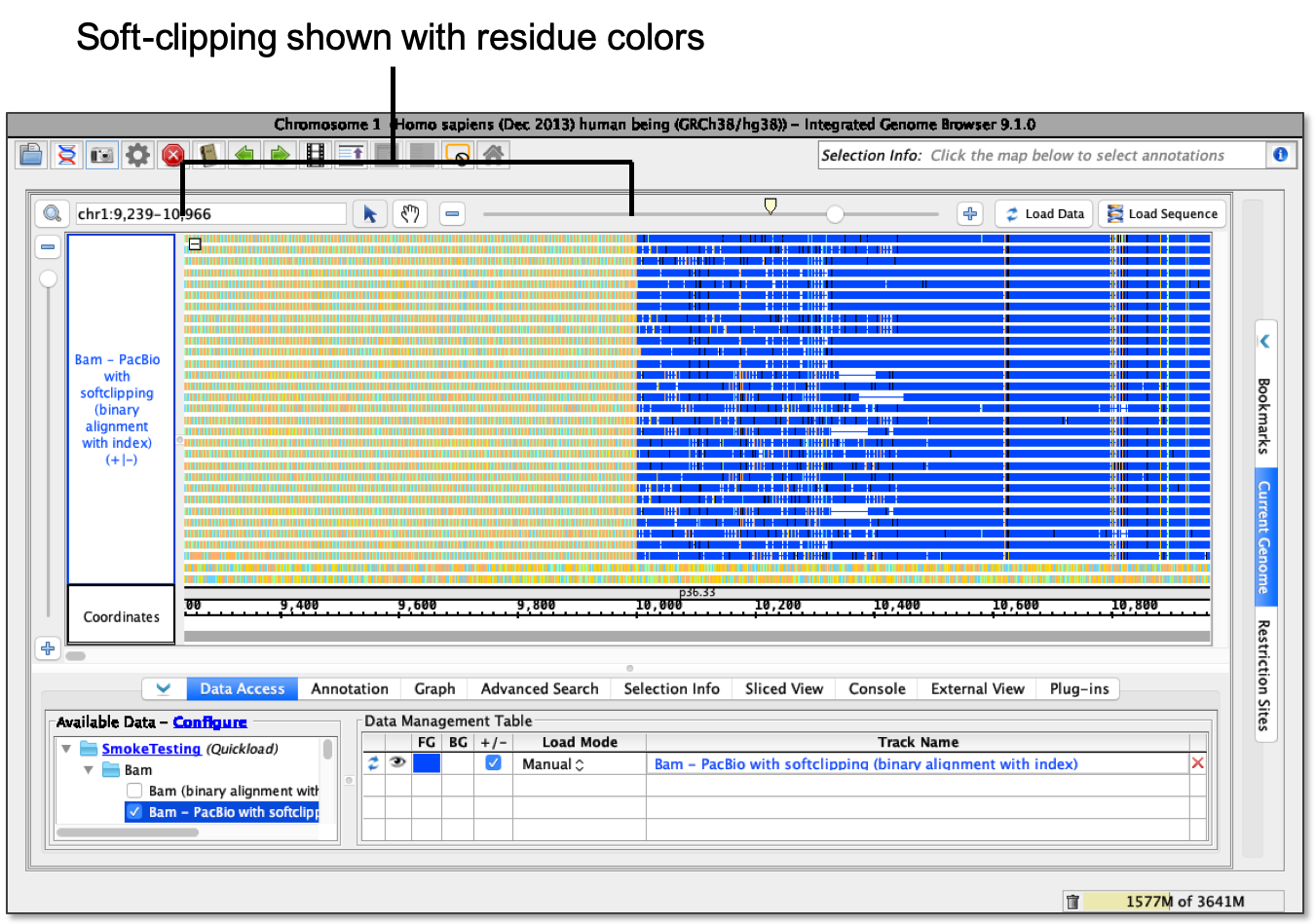
- Show as bases - softSoft-clipping shows as Residue Colors.
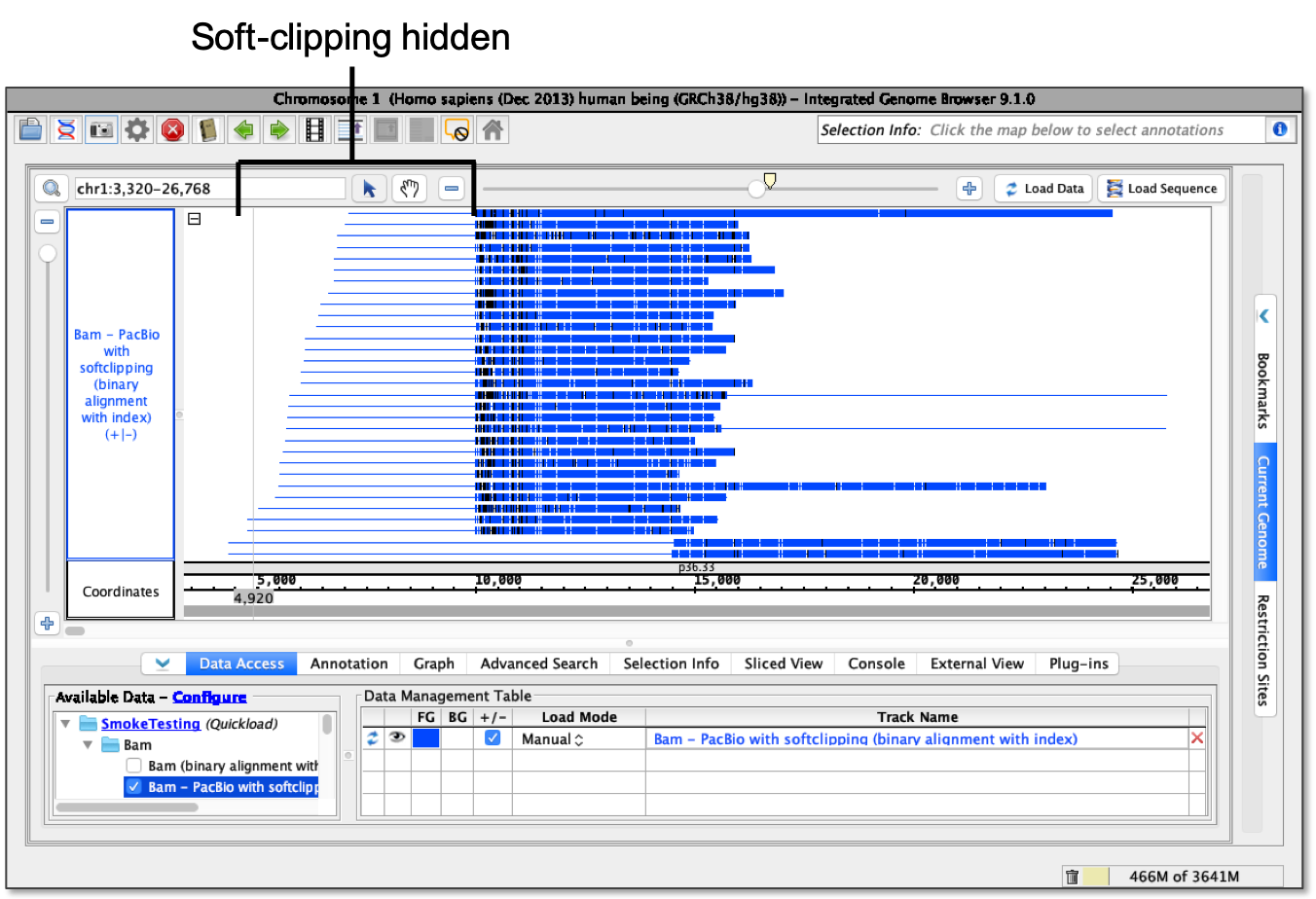
- Hide soft-clipping - softSoft-clipping is hidden.
Note: When using the hide soft-clipping option, a line showing the full length of the read including the soft-clipped portion will appear.
To completely hide the soft-clipping portion of the read, use the Show as Custom Color soft-clip option, and select the same color as the background for the track.
Track Operations and Soft-clipping
Track Operations behave differently when soft-clipping is present.
- Depth Graph (All) and Depth Graph (Start) ignore the soft-clipped parts of reads.
- Mismatch Graph and Mismatch Pileup Graph include the soft-clipped parts of reads.
Data used in these examples is a subset taken from the Genome in a Bottle consortium (HG002 PacBio CCS 10kb):
Zook JM, Catoe D, McDaniel J, et al. Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci Data. 2016;3:160025. Published 2016 Jun 7. doi:10.1038/sdata.2016.25