...
Mismatch Graph
The Mismatch graph Graph is specific to short read alignment files, such as BAM. The graph shows only the number of mismatched nucleotides across all reads at a specific genomic location, which can be very helpful in the detection of allelic variation, SNP identification and also error checking. Right click in the label to access the menu; choose Track Operations > Mismatch Graph. This function requires that the genomic sequence be loaded; if it is not loaded, IGB will load the sequence for you.
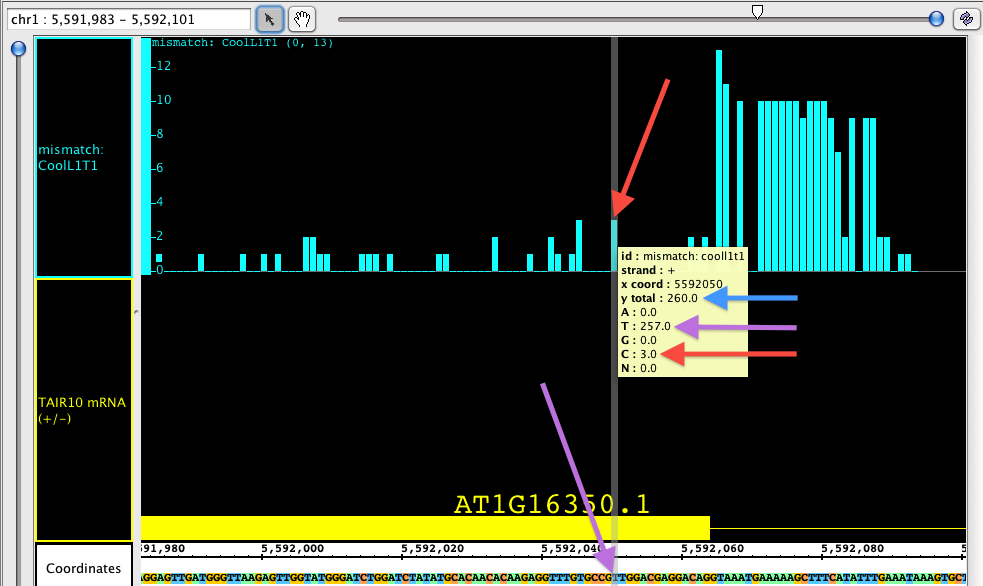
If you hover the Select tool (arrow cursor) over an individual bar of the graph, you will get a tool tip showing the total number of reads, and then the number of reads broken down by nucleotide. In the picture below, you can see that the total number of reads at the specified position is 260 (blue arrow). Of those reads, 257 at at 'T', which you can see is the matching nucleotide (purple arrows). 3 of those reads contain a 'G' instead; since G is a mismatch, the graph shows a bar with height of 3 (red arrows).
Mismatch Pileup Graph
In the Mismatch Pileup, you have the Depth portion represented as a dark gray graph, with the Mismatch overlayed in the matching nucleotide colors. The colors show which and how many of each nucleotide(s) are present at each mismatch position. The Mismatch Pileup graph, like the Mismatch graph, is also specific to short read alignment files, such as BAM. Right click in the label to access the menu; choose Track Operations > Mismatch Pileup Graph. This function requires that the genomic sequence be loaded; if it is not loaded, IGB will load the sequence for you.
...