Insertions
Insertions appear as a pair of red residues. The location of the insertion lies between these red residues. Details about the insertion can be found in the Selection Info tab or by hovering the mouse over the red residues to activate the tooltip. The image below shows a single residue insertion of 'C' that lies between the residues 'CT'.
Deletions
Dash characters on a gray background represent deletions in the aligned sequence with respect to the genomic sequence. In other words, a dash character represents one or more contiguous bases that were present in the reference genomic sequence and absent from the aligned sequence.
Mismatches
Mismatches between the reads and the reference sequence are highlighted according to the mismatched base. In the image below, all five reads have a mismatch to A, indicating a single nucleotide polymorphism. One read has two mismatches to G, but these mismatches are not supported by any other reads.
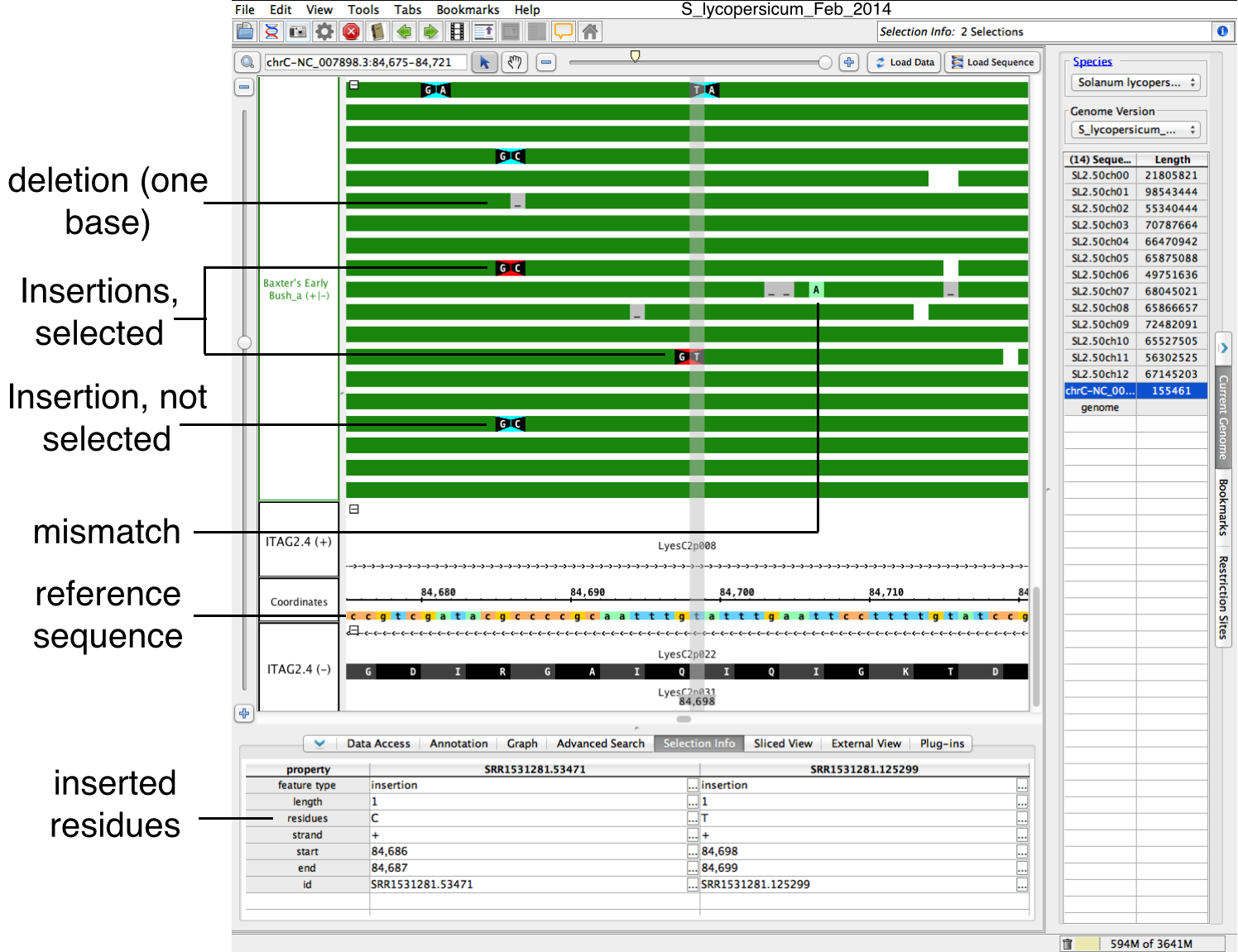
IGB displays insertions, deletions and mismatches as color-coded bases and icons on top of read alignments, as shown in the following image.
- Insertions - bases flanking insertion site appear are shown in white with a black background; triangular shapes between them point at the location of the inserted base or bases that are present in the aligned sequence but not in the reference. Select an insertion to display the sequence of inserted bases.
- Deletions - bases deleted in the aligned sequenced relative to the reference appear in gray with underscore characters
- Mismatches - bases in the aligned sequence that do match the corresponding base in the reference appear as higlighted bases. To highlight mismatches, clickLoad Sequence.