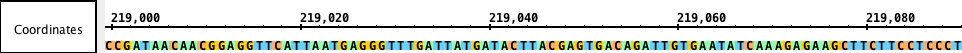Genomes and chromosomes
Chromosome, also called contig or reference sequence, refers to one or more annotated sequences that form a genome assembly. For completed genome projects, these often correspond to the sequence of a physical chromosome. Alternatively, they may represent assembled contigs corresponding to parts of a physical chromosome.
Genome, genome version, or assembly refer to a group of annotated sequences corresponding to the genome sequence of an organism. IGB designates these using the month and year they were published or made publicly available.
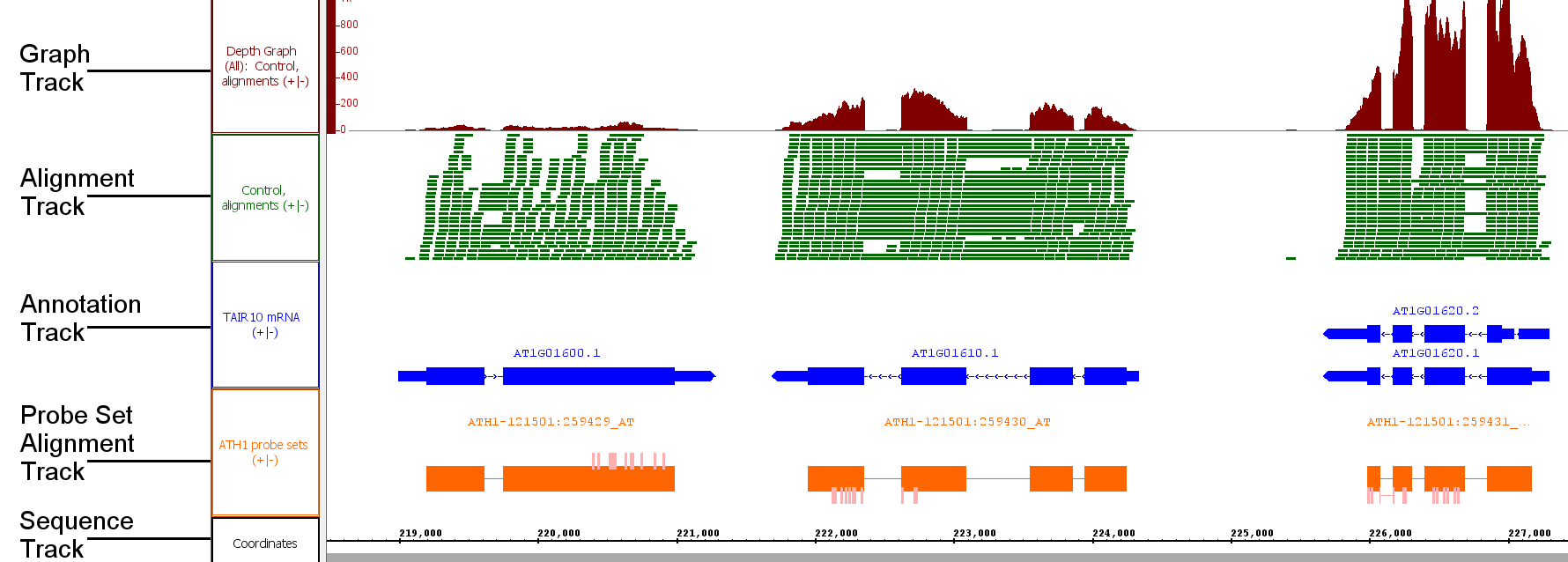
Tracks
Tracks are rows of data that appear in the IGB interface. When you open a file, the data within the file will appear in one or more tracks. Older versions of IGB refered to tracks as "tiers" and so you may see this term used elsewhere in the User's guide.
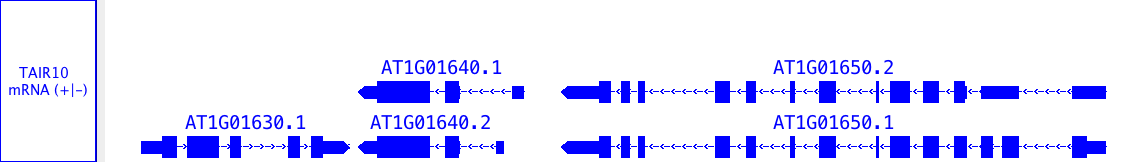
Annotation tracks
Annotations indicate the known or suspected locations of genomic features such as genes, exons, promoter regions, pseudogenes, and so forth. Annotations may consist of a single coordinate, a single span with a start and end positions, or a collection of spans. Most annotations reside on either the plus or minus strand of a chromosome, but some do not.
Examples of annotations include:
- single-coordinate feature: splice sites
- single-span feature, with strand: an exon
- single-span feature, no strand: a restriction site
- multi-span feature, with strand: a gene model
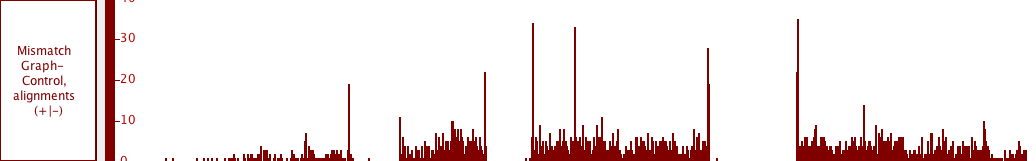
Graphs tracks
Graphs are numeric data associated with regions or single-base positions in a chromosome.
Sequence track
Sequences are DNA residues from a chromosomes or contigs that together make an assembly. Sequences can be fully or partially loaded from local files, Quickload sites, or Distributed Annotation Servers. You can view sequence data in the Coordinates track or by opening the Sequence Viewer.
Alignments
Alignments represent how sequences obtained from an experiment (such as reads from an RNA-Seq experiment) align onto a reference sequence. At low zoom they look like annotations, with marks representing mismatches, insertions, or deletions. At higher zoom, the aligned sequence bases become visible.
Probe set alignments
Probe set alignments consist of probe set target sequences aligned onto the reference with probe locations indicated as annotations on the target sequence alignments. The probes are kind of annotation on an annotation.
These data are Affymetrix specific.