General Function Checklist
Objective: Validate that soft-clipping can be hidden and customized using the right-click menu of a bam track, as shown below:
- In the Data Sources tab in Preferences, add the smoke test Data Provider to IGB using the following URL: https://quickload-testing.s3.amazonaws.com/smokeTestingQuickload/
- Open the H_Sapiens_Dec_2013 genome.
- In the Available Data panel, add the Bam - PacBio with softclipping data from your newly added Quickload.
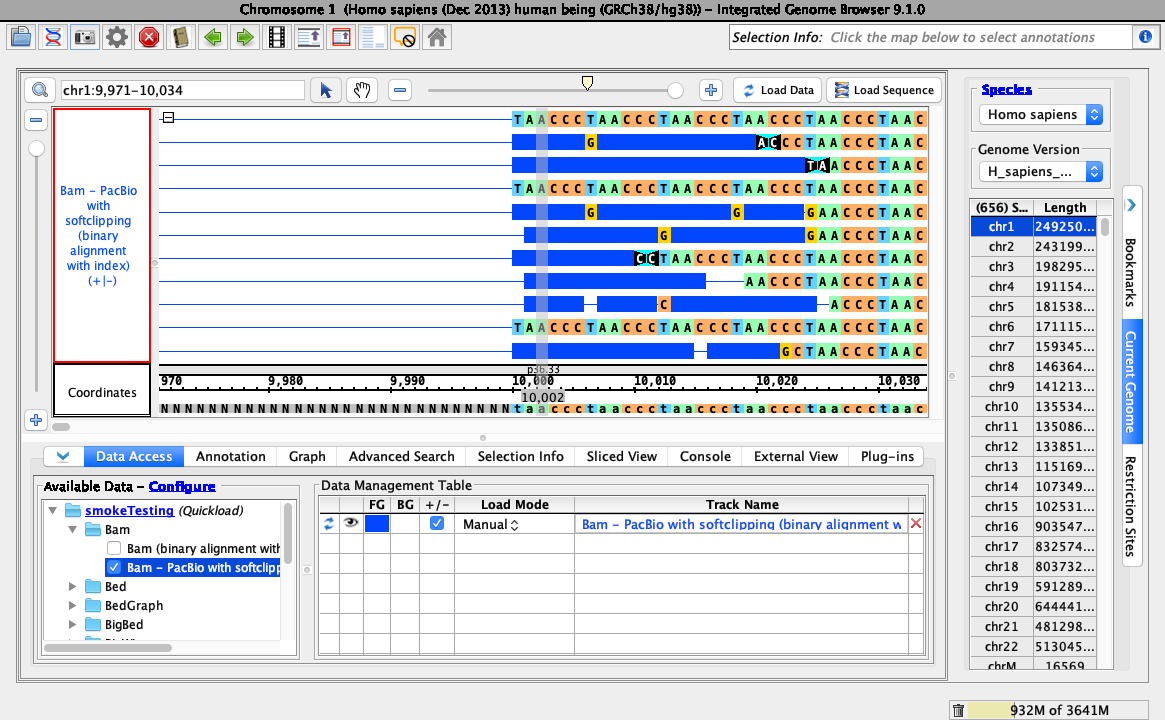
- Navigate to: chr1:9,971-10,034
- Click Load Sequence to load sequence data, then click Load Data to load data into it.
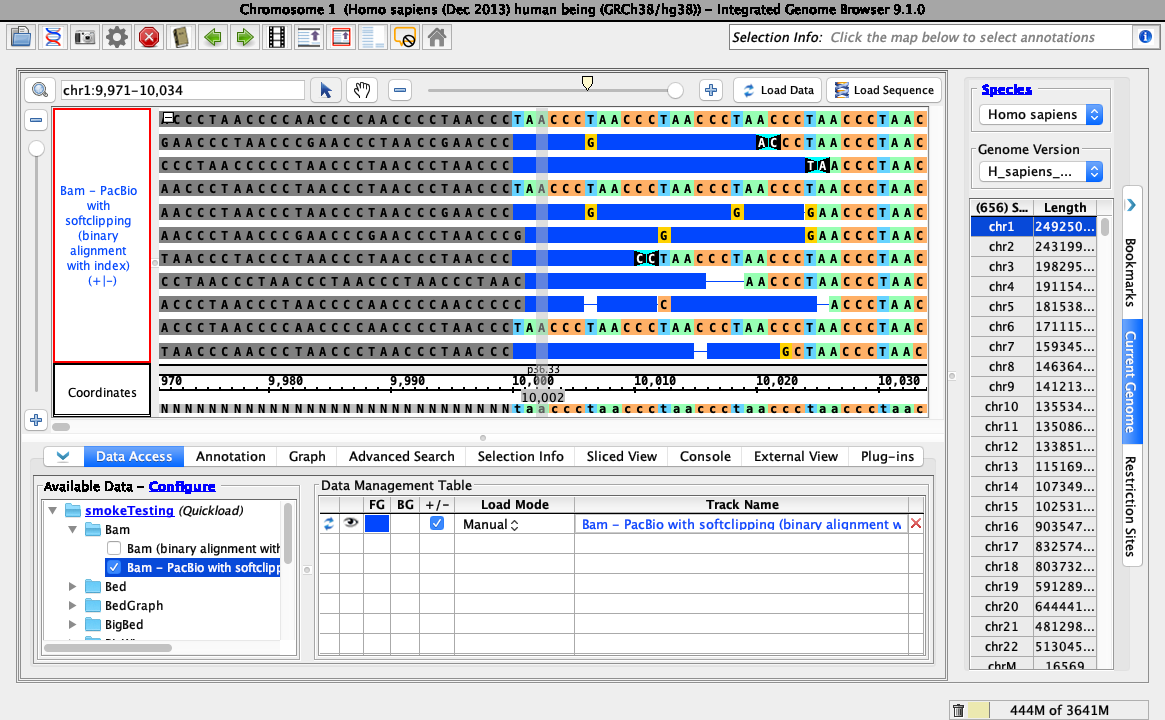
- Loading data looks like this (stack height set at 10, reads may be organized randomly):
- mac
- linux
- windows
- The Configure soft-clip submenu of the bam track right-click menu has Show as default color selected.
- mac
- linux
- windows
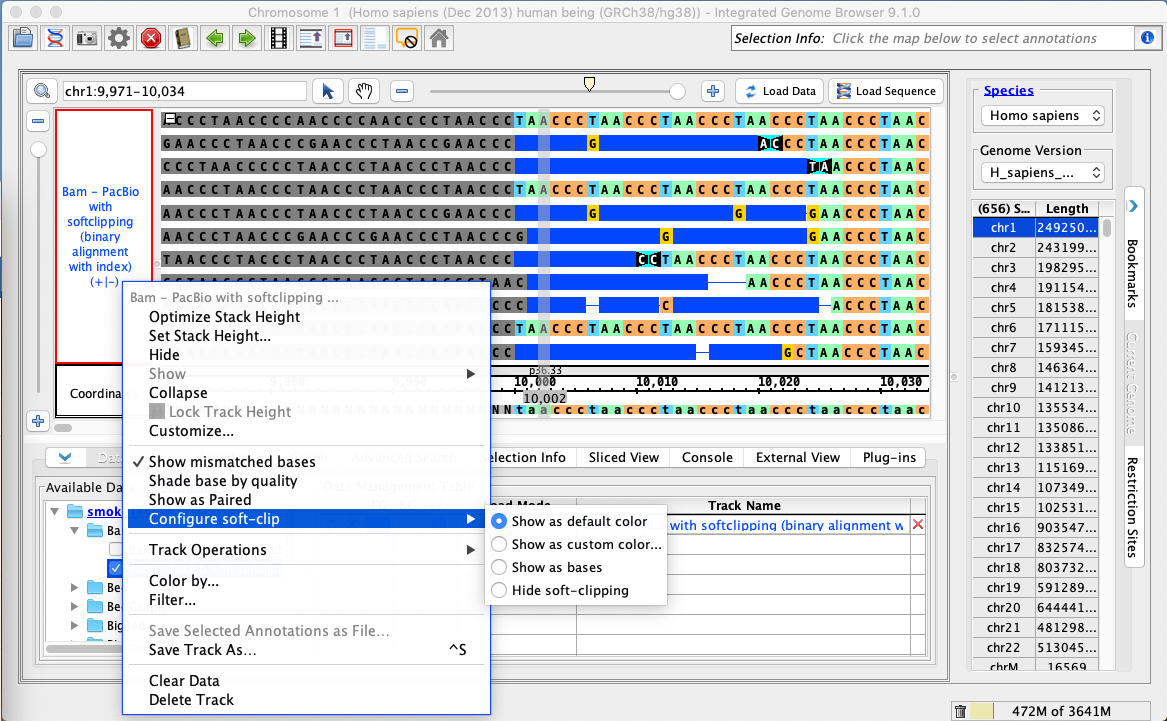
- Right-click on the bam track and select: Configure soft-clip -> Show as custom color.
- Choose a red color from the color picker.
- Your IGB looks like the above image.
- mac
- linux
- windows
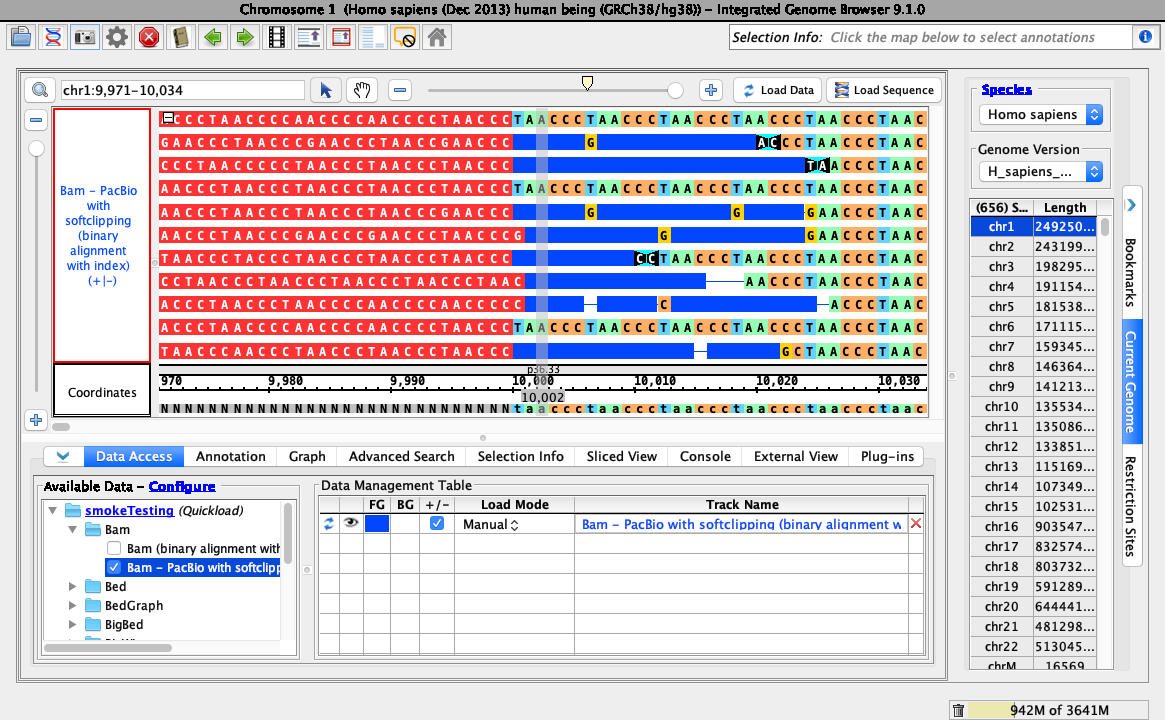
Right-click on the bam track and select: Configure soft-clip -> Show as bases.
- Your IGB looks like the above image.
- mac
- linux
- windows
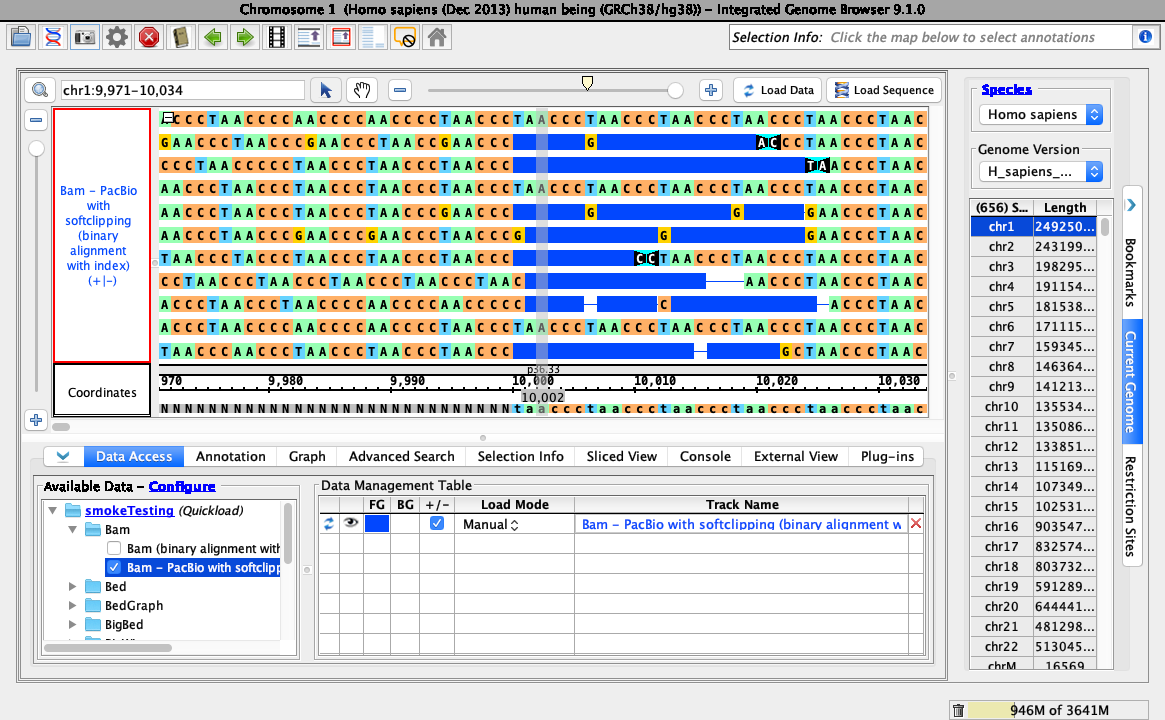
Right-click on the bam track and select: Configure soft-clip -> Hide softclipping.
- Your IGB looks like the above image.
- mac
- linux
- windows
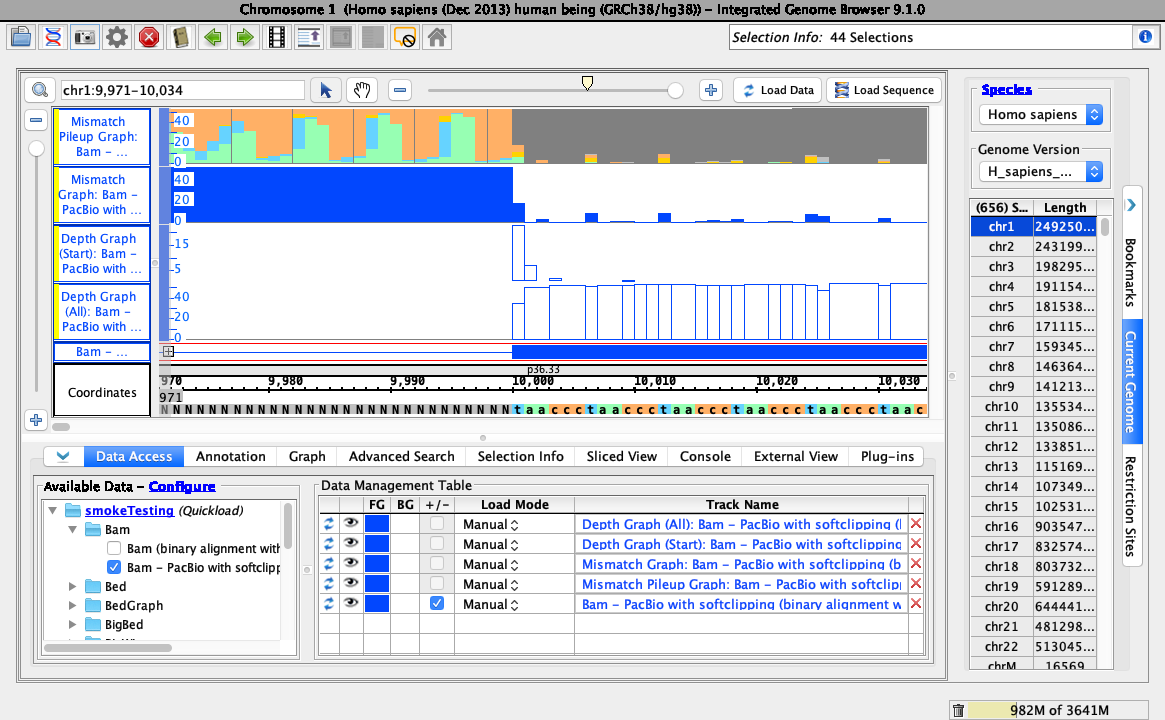
To test track operations are working correctly for soft-clipped data, right-click on the bam track and select the following:
- Track Operations > Depth Graph (All)
- Track Operations > Depth Graph (Start)
- Track Operations > Mismatch Graph
- Track Operations > Mismatch Pileup Graph
- Collapse
- Your IGB looks like the above image.
- mac
- linux
- windows