A common analysis step is to determine meaningful regions on a sequence based on graph values being above or below a certain threshold. To screen out non-meaningful values and to view meaningful features of a graph as annotations, use the thresholding feature in IGB.
Thresholding displays graph features as annotation-like bars in locations where the graph value at the coordinate meets your defined threshold. You can convert this data into a standard annotation track in order to analyze your graph data using all of the features in IGB for working with and comparing annotations.
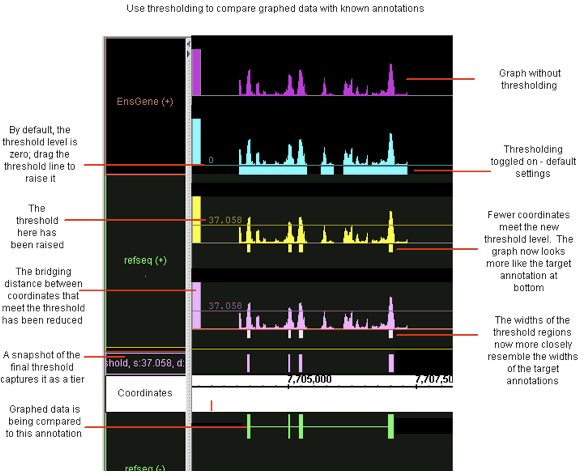
In some cases, you may want to adjust your graph thresholds so the bars correlate to existing annotations; any threshold bars visible at that point that don't have known annotation counterparts (or vice-versa) may indicate areas for further investigation.

To use thresholding:

To change the thresholds of selected graphs so that more or fewer points meet the threshold limit, modify these settings in the Graph Thresholds window:
Specify whether the threshold bars represent values that are above the threshold you specified or below it.
In the Thresholding section of the Graph Adjuster tab, choose:
For example, for expression results graph score represents the level of hybridization activity. For a hybridization activity level to be considered significant, the score must be above a certain threshold. So you would choose the > thresh option and set your desired threshold value. If you need help to pick a threshold level, toggle the Y-axis on to see the graph's scale on the Y-axis. See Graph labeling on page 57.
Experimental data can be noisy. If you are looking for general trends and want to ignore small local variations, you can use the Max Gap and Min run settings.
Slide the sliders to adjust thresholding, or enter values into the boxes for each parameter:
The Offsets for Thresholded Regions feature is designed specifically for working with tiling array data.
Captured data from tiling array probes should usually be shifted for display. Since the probes are 25 base pairs long, but the x-coordinates given represent the starting coordinate, you should shift the threshold data so that it starts at 12 base pairs past the given beginning, and ends at 13 base pairs past the beginning. This is the default placement of all graph threshold bars.
If these offsets are not correct for the data you are analyzing, you should change them.
If you are viewing typical tiling array data, modify these settings in the Graph Thresholds window:
When you view non-tiling array data in IGB, you should adjust the offset to 0 to correctly align the threshold bars with their actual coordinates.
To adjust the offset values for non-tiling array data modify these settings in the Graph Thresholds window:
Capture the threshold bars (which look similar to annotations) in their own track to more easily compare different threshold settings for a single graphed region. (See the thresholding illustration, above)
To capture the threshold bars and convert them to annotation tracks:
A snapshot of the current thresholded graph will be created as a new annotation track. This track will be given a default label which you can later change if desired.
To permanently capture the threshold snapshot for later use or sharing:
After you have captured threshold "annotations" from a graph or other set of data, you can examine these "annotations" using all of the tools in IGB for working with and comparing annotations.