
All Default DataProviders appear in the DataProviderManagementGui table

Confirm that clicking on a [i] icon in the rightmost 'information' column of the Data Sources window above opens a window with fields and values similar to the example below:
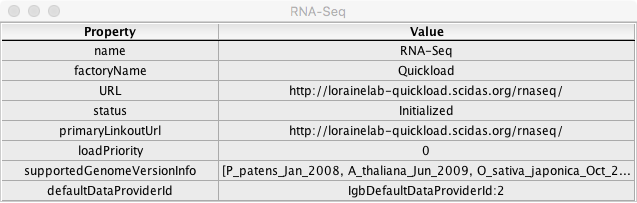
Confirm that cells in the above information window contain information related to their respective data source
Confirm that closing this window and opening another shows correctly updated information
Confirm that the information window functions for each data source type
Confirm the following species / genome versions are available
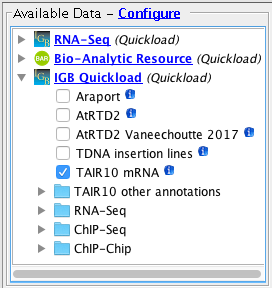
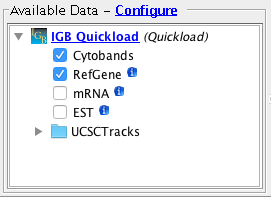
Important: Confirm that for the human genome assembly, the Cytobands track is visible

Bed File
Bam File
Bedgraph File
Load Sequence
Adding/Removing/Editing DataProviders
Add a quickload data provider and confirm it is working as expected
Enter a name for this quickload site and click the "Submit" buttonObservations:
Confirm restarting IGB does not cause the newly added site to be forgotten
Ensure that the edit button in the Data Sources window opens a new window and allows a user to modify the data source URL or to choose a local folder. Note that default data sources cannot be modified.
Ensure that data sources can be deleted using the Remove button in the Data Sources window
Add a few "Secured" quickload sites and confirm they are working as expected
http://igbquickload.org/secureQuickloadTestSites/secureSiteTest
username/password
http://igbquickload.org/secureQuickloadTestSites/secureSiteTest2
username/password
Navigate to the Arabidopsis thaliana Jun_2009 genome version
Confirm the two sites are listed the "Available Data" tree and data can be loaded from each site
Confirm you were not prompted for a password for any site more than a single time per session
Confirm restarting IGB does not cause the newly added sites to be forgotten
Confirm that if you checked the "Save Password" option you are not prompted for your password again on restarting IGB