General Function Checklist
Verify that all commonly used file formats are properly supported in IGB by loading all files available on the Smoke Testing Quickload.
- Add a Data Provider to IGB using the following URL: http://igbquickload.org/smokeTestingQuickload/
- The added Quickload site is available for the Homo sapien genome under the Available Data pane.
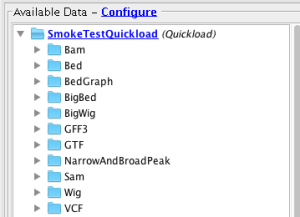
- Verify that all files in the Quickload site are loading into IGB:
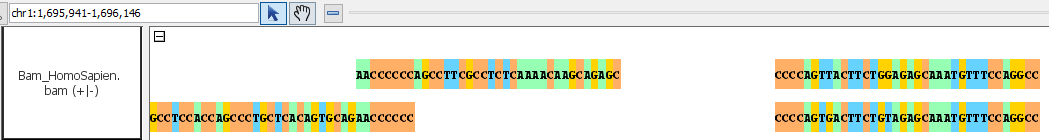
- Bam
- Bam_HomoSapien.bam
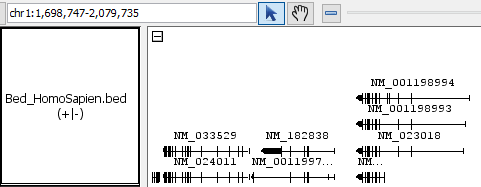
- Bed
- Bed_HomoSapien.bed
- Bed_HomoSapien.bed.gz
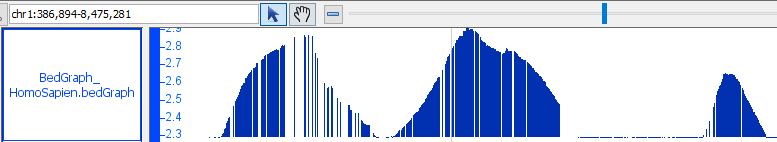
- BedGraph
- BedGraph_HomoSapien.bedgraph
- BedGraph_HomoSapien.bedgraph.gz
- BigBed
- BigBed_HomoSapien.bigbed
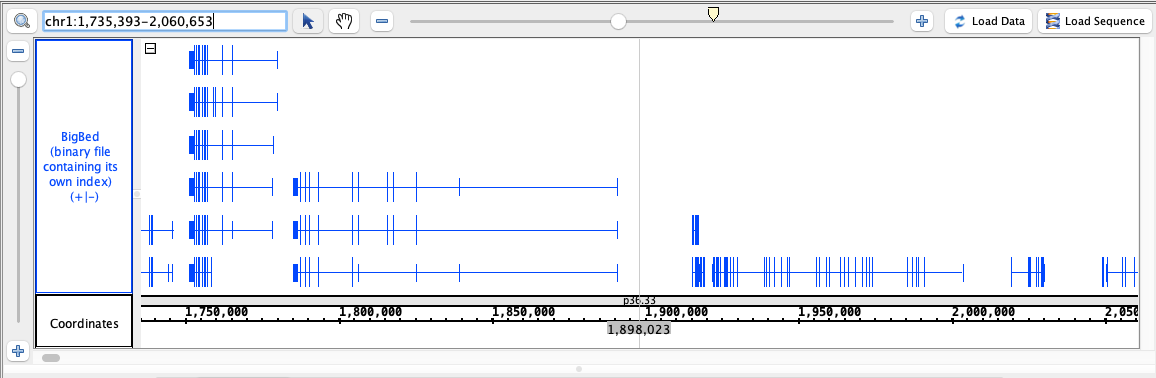
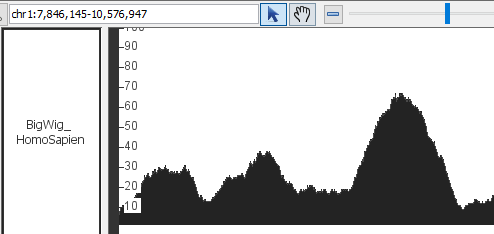
- BigWig
- BigWig_HomoSapien,bigwig
- GFF3
- GFF3_HomoSapien.gff3
- GFF3_HomoSapien.gff3.gz
- GTF
- GTF_HomoSapien.gtf
- GTF_HomoSapien.gtf.gz
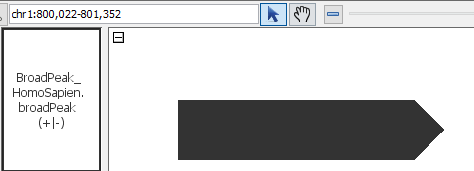
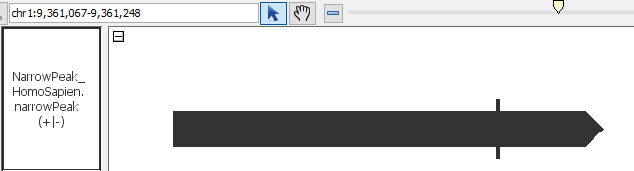
- NarrowAndBroadPeak
- BroadPeak_HomoSapien.broadpeak
- BroadPeak_HomoSapien.broadpeak.gz
- NarrowPeak_HomoSapien.narrowpeak
- NarrowPeak_HomoSapien.narrowpeak.gz
- VCF
- VCF_HomoSapien_Mutations(SNPs).vcf
- VCF_HomoSapien_Mutations(SNPs).vcf.gz
- Can you see mutations (SNPs)?
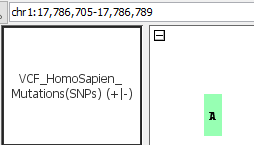
- VCF_HomoSapien_InsertionsAndDeletions.vcf
- VCF_HomoSapien_InsertionsAndDeletions.vcf.gz
- Can you see deletions?
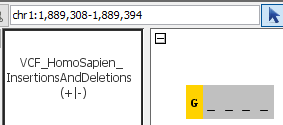
- Can you see insertions?
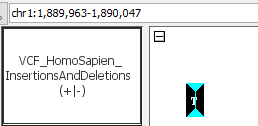
- Wig
- Wig_HomoSapien.wig
- Wig_HomoSapien.wig.gz
Sequence appearance
Go to the Arabidopsis genome. From the RNA-Seq quicklaod (available by default as of 9.0.1), open the file RNA-Seq / Pollen SRP022162 / Reads / Pollen alignments. (url http://lorainelab-quickload.scidas.org/rnaseq/A_thaliana_Jun_2009/SRP022162/Pollen.bam)
Go to location: Chr1:7,319,301-7,319,375
Optimize the stack height, and compare the main view to this image:
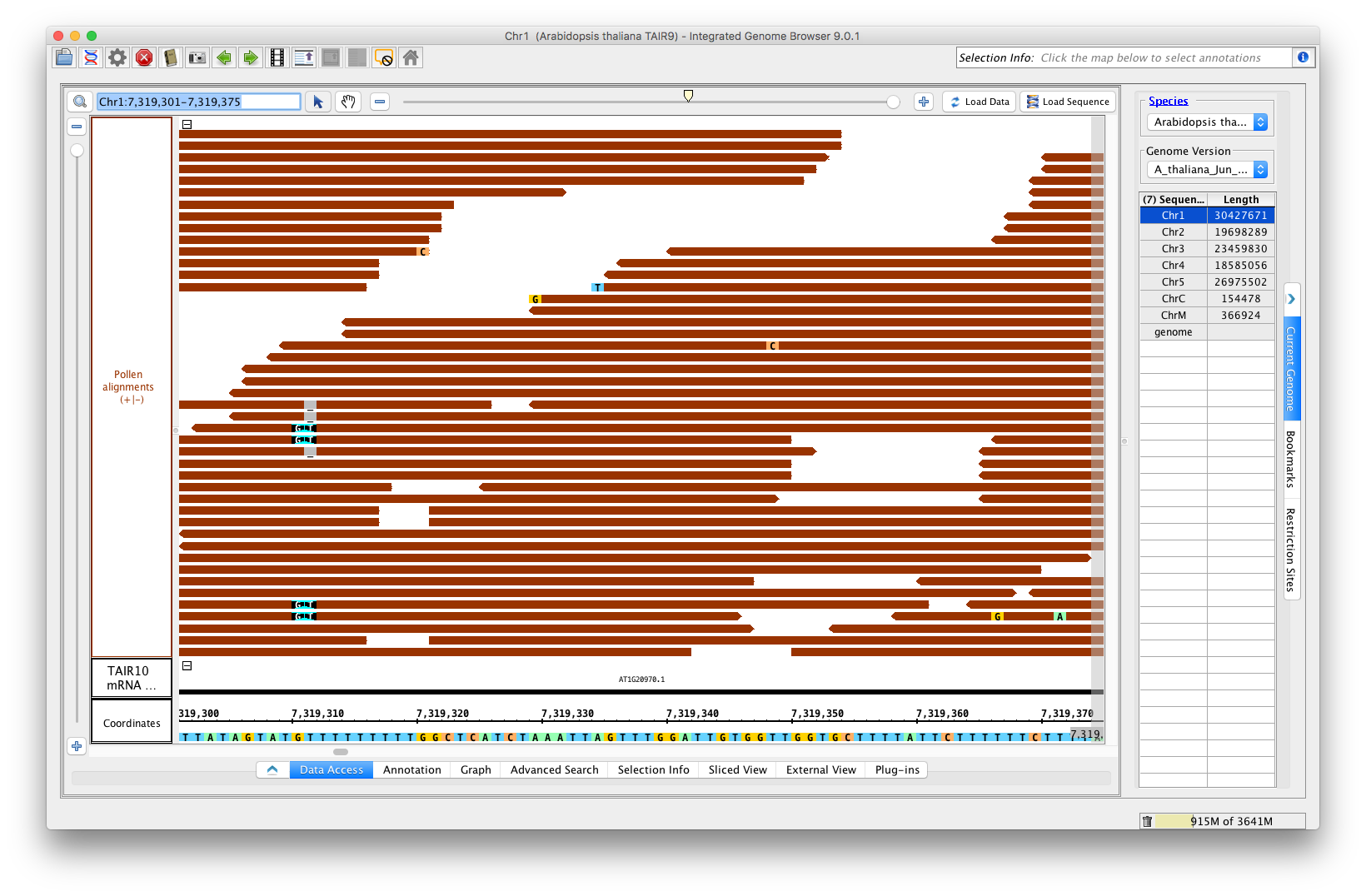
Verify that:
- single base mismatches of A, T, C, and G appear with a color scheme that matches the sequence in the coordinate axis (the bottom).
- deletions appear as shown, a gray space with a dash.
- insertions appear as shown (see position 7,319,310 in image)











