Be sure to look at the user guide pages linked here. Skim them to make sure that the topics they cover are represent here (if not add points here as needed). Read enough to ensure that the instructions and explanations are clear, and the page has accurate information and generally appears up-to-date.
Annotation Track Operators
See Users Guide pages:
single track - output an annotation track
Go to the Arabidopsis thaliana June 2009 genome.
Go to this region: Chr1:2,992,975-3,012,448
Load the TAIR10 mRNA (can be found in the 'TAIR10 other annotations' folder).
Select the annotation tab, and look for the Operations box.
- copy (single track)
- Select the ( - ) track and select copy, hit go
- Select the ( + ) track, select copy, hit go.
- combine the ( - ) and ( + ) tracks into one using the (+/-) checkbox.
- Select the (+/-) track, select copy, hit go.
- In the annotation tab, use Select All, and under Strand, check the Arrow option (to make visual verification easier)
- Verify that the tracks produced match this image: (matching the colors is optional)
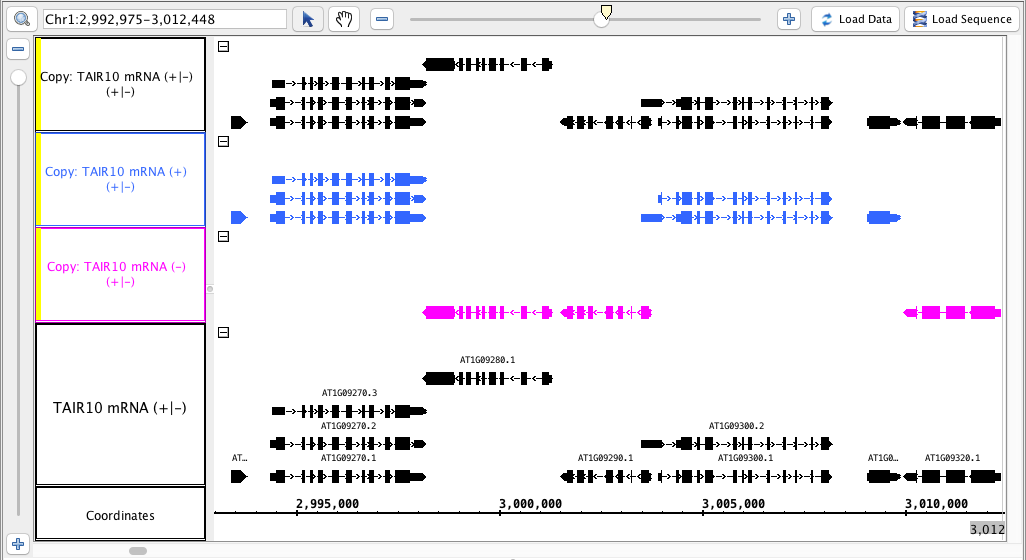
- the track names for the new tracks indicate which track they were made from AND which strand ( -, + or +/- )
- Mac
- Linux
- Windows
The tracks made from only one strand only have the annotations from that strand. - Mac
- Linux
- Windows
The track made from both has annotations from both strands. - Mac
- Linux
- Windows
Delete the tracks you just created. Split the mRNA track back into separate - and + tracks.
- Not (single track)
- Select the ( - ) track, select Not, hit Go.
- Select the ( + ) track, select Not, hit Go.
- Recombine the - and + tracks.
- Select the ( +/- ) track, select Not, hit Go.
- Summary (single track)
- Repeat all of the above for the summary operation.
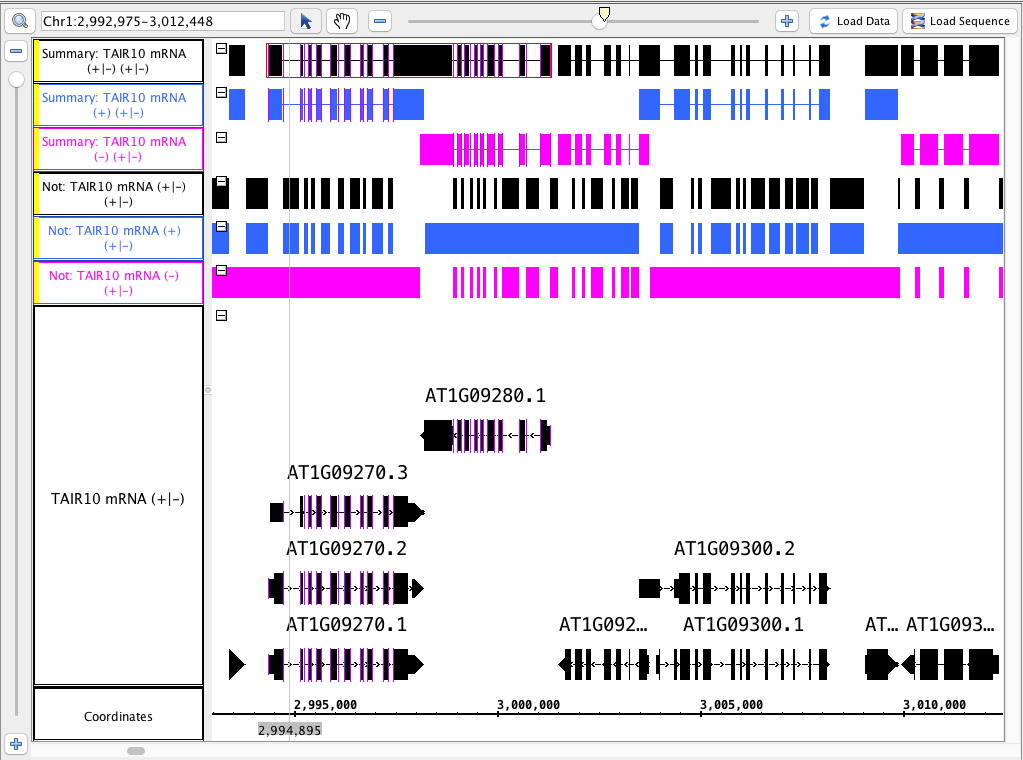
- Color the tracks that were made from the ( - ) strand pink and the ( + ) blue. (for visual verification)
- Compare the results to this image below (or logic through it):
multi track (output annotation track)
- Using the combined (+/-) mRNA track, run the Summary operation and the Not operation.
- Hide the original track and highlight the two newly created tracks (hold shift and click each one) --Select the "Not" track first.
- Perform each of the multi-track operations:
- Compare the results to this image below (or logic through it):
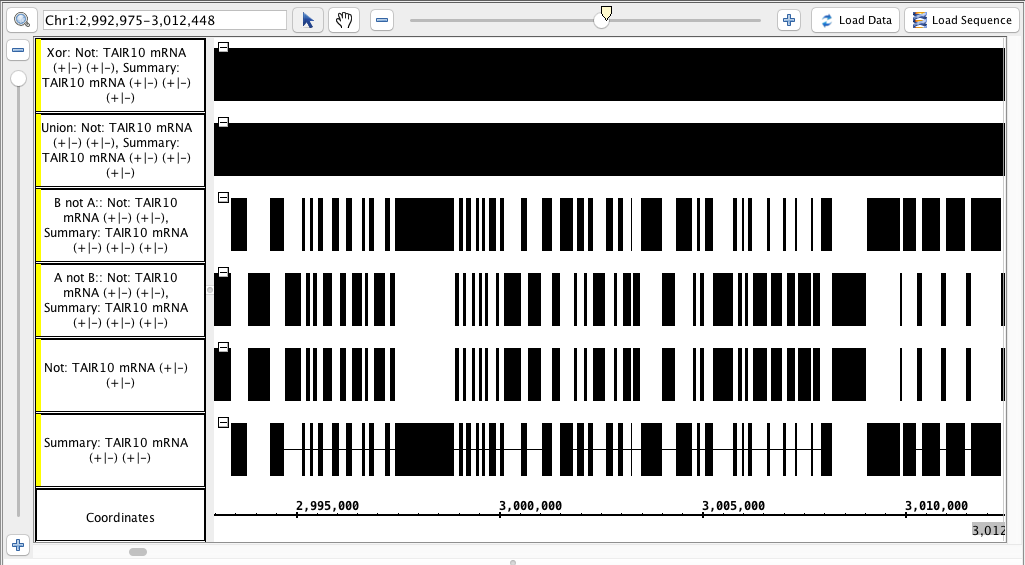
- Track labels include the name of the operation and the input tracks
- Visually inspect the track is identical to the "Not" track, or identical to the "Summary" track, or completely solid, as appropriate.
- Delete the tracks you just created.
- Using the split ( - ) and ( + ) mRNA track, perform each of the multi-track operations:
- Compare the results to this image below (or logic through it):
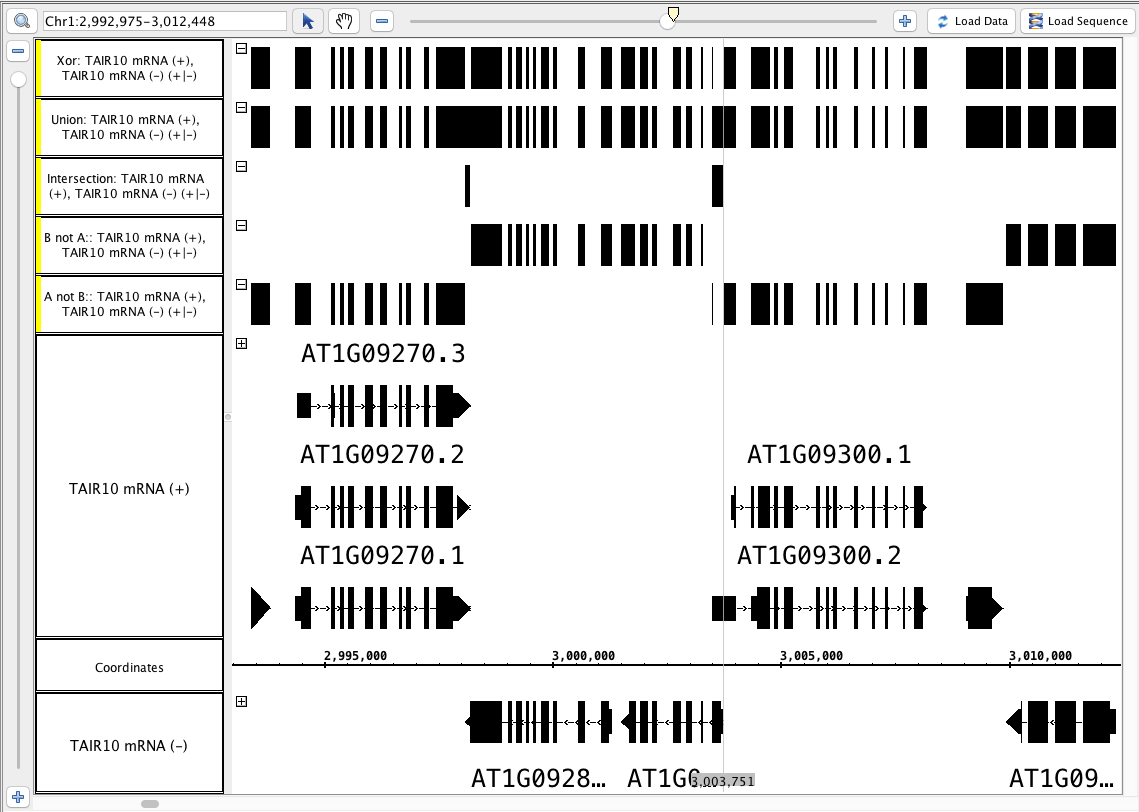
- Particularly notice the area around the zoom stripe, it highlights the difference between Union and Xor.
single track - output a graph track from a bed file
Keep using the Arabidopsis thaliana June 2009 genome annotations file, keep using this region: Chr1:2,992,975-3,012,448
- Use the + and - tracks separately to run each of these operations.
- depth (all) (single track)
- depth (start) (single track)
- This produces 4 graph tracks.
- Repeat this by selecting both the - and the + track at the same time (hold shift) and select these single track operations.
- The results should be identical
- Select one of the depth all graphs. In the graph panel, Choose Thresholding... . In the thresholding window:
- Slide the By Value slider
- observe the little blocks appear and disappear as the line touches the graph.
- verify that the values in the graph track correspond to counts of annotations that cover each location
- Choose Make Track
- Verify that the new track looks correct.
- Verify that the new track has an appropriate label (that indicates what threshold was used to create it)
- See image
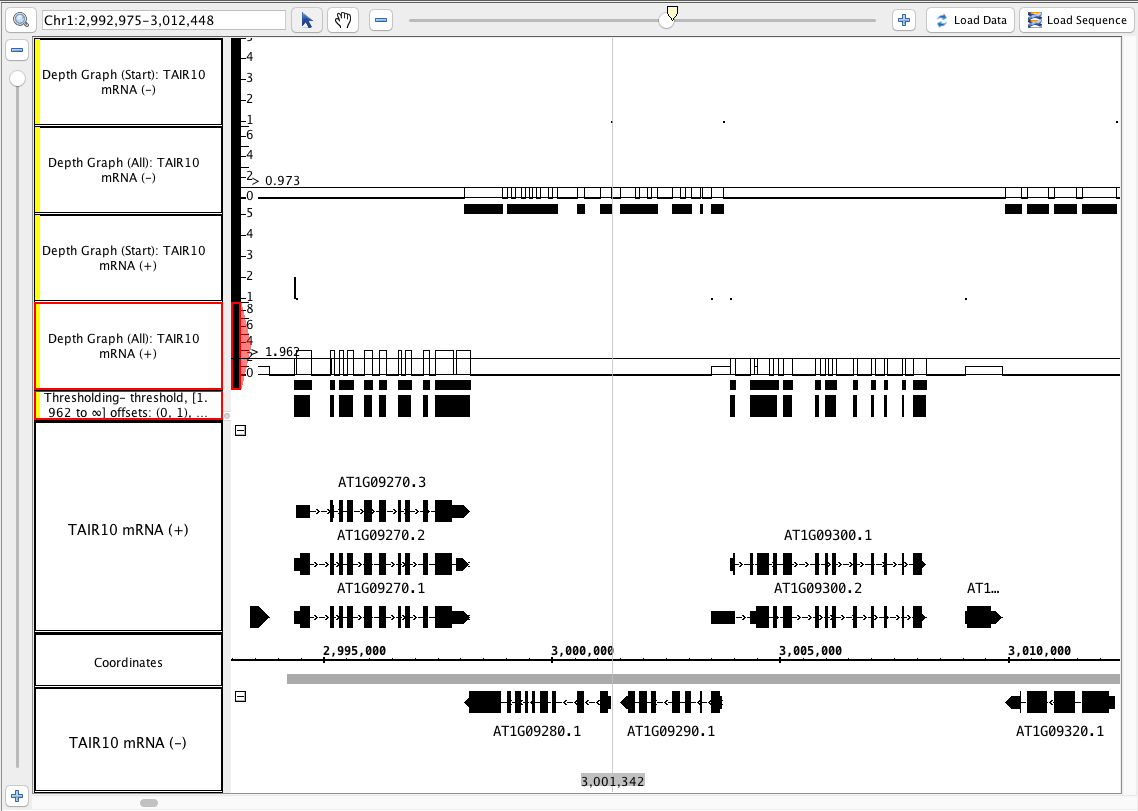
- notice that the depth (start) graphs have data in a 1-base-wide area at the start of the annotations, and where there is only 1 annotation, the graph has a height of 1.
single track - output a graph track from a bam file (part 1)
Keep using the Arabidopsis thaliana June 2009 genome annotations file. Go to this region: Chr1:2,998,442-3,001,453
From the RNA-Seq quickload, add the RNA-Seq / Pollen SRP022162 / Reads / Pollen alignments file.
Click Load Data
Click Load Sequence
Select the Pollen alignments track and run the following single track operations:
- Select Find Junctions, enter 5, hit go.
- Select Find Junctions, enter 10, hit go.
- Select Find Junctions (tophat), enter 5, hit go.
- Select Find Junctions (tophat), enter 10, hit go.
Compare to this image below:

* Read the junctions user guide page for more details.
- Check the sizes of the blocks for the Find Junctions tracks (it should match the value you entered); the find junctions (tophat) results will not all have the same block size.
(Right click track > optimize track height. This will show all of the reads.)
- Verify that the count above each junction is equal to the number of reads that matched that gap.
- Find someplace where the matching Find Junctions operation using value 5 and value 10 have different scores. Identify the read(s) that matches that junction, and where where one side of the read is between 5 and 10 bases.
single track - output a graph track from a bam file (part 2)
Go to the human genome. Add the smoke testing quickload: http://igbquickload.org/smokeTestingQuickload/. Add the bam file.
Go to this region: chr1:2,180,492-2,180,751
Load data, Load Sequence.
Select the Bam_HomoSapian track and do the following single track operations:
- depth (all) (single track)
- depth (start) (single track)
Mismatch Graph
Mismatch Pileup Graph
Compare the results to this image (and look at them to see that they make sense!)
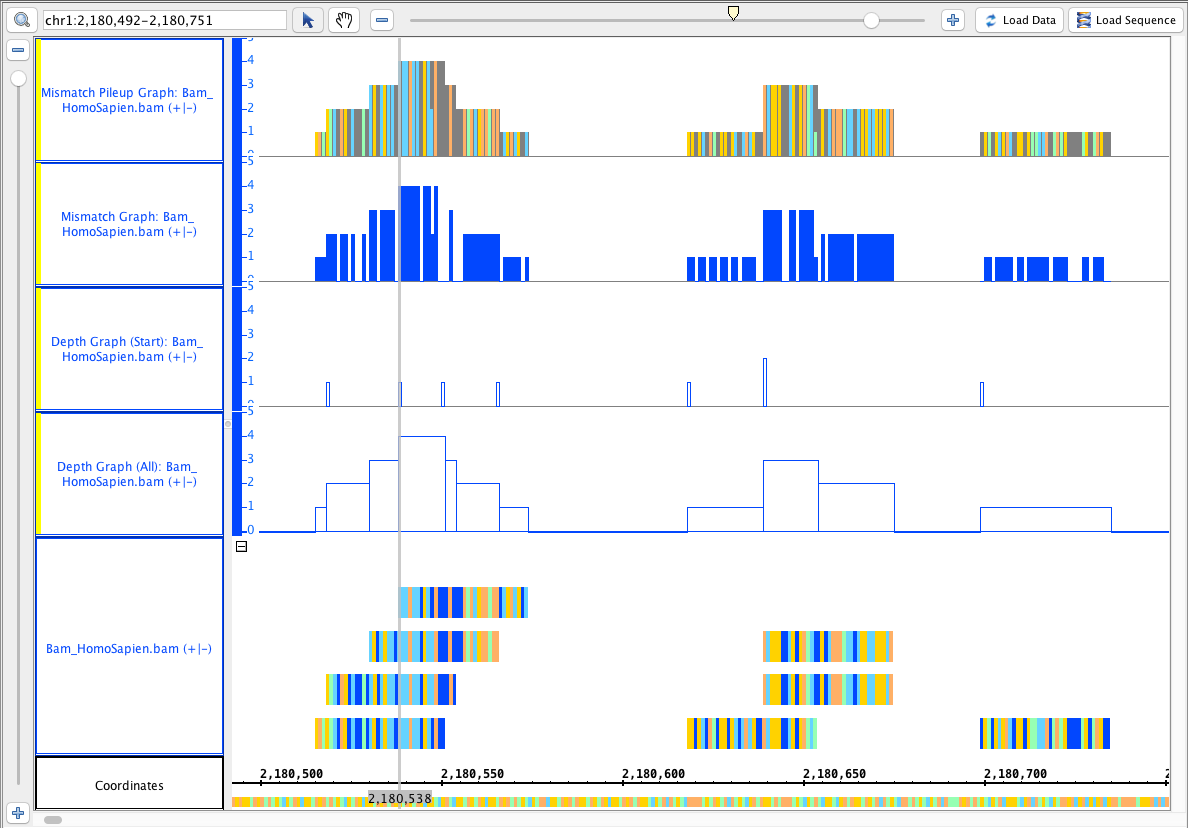
Zoom out. Load data (do not load sequence). Load data a second time if needed, but not a third.
- verify that the sequence is loaded as needed.
Go to this region: chr1:3,785,135-3,785,276, it should look like this:
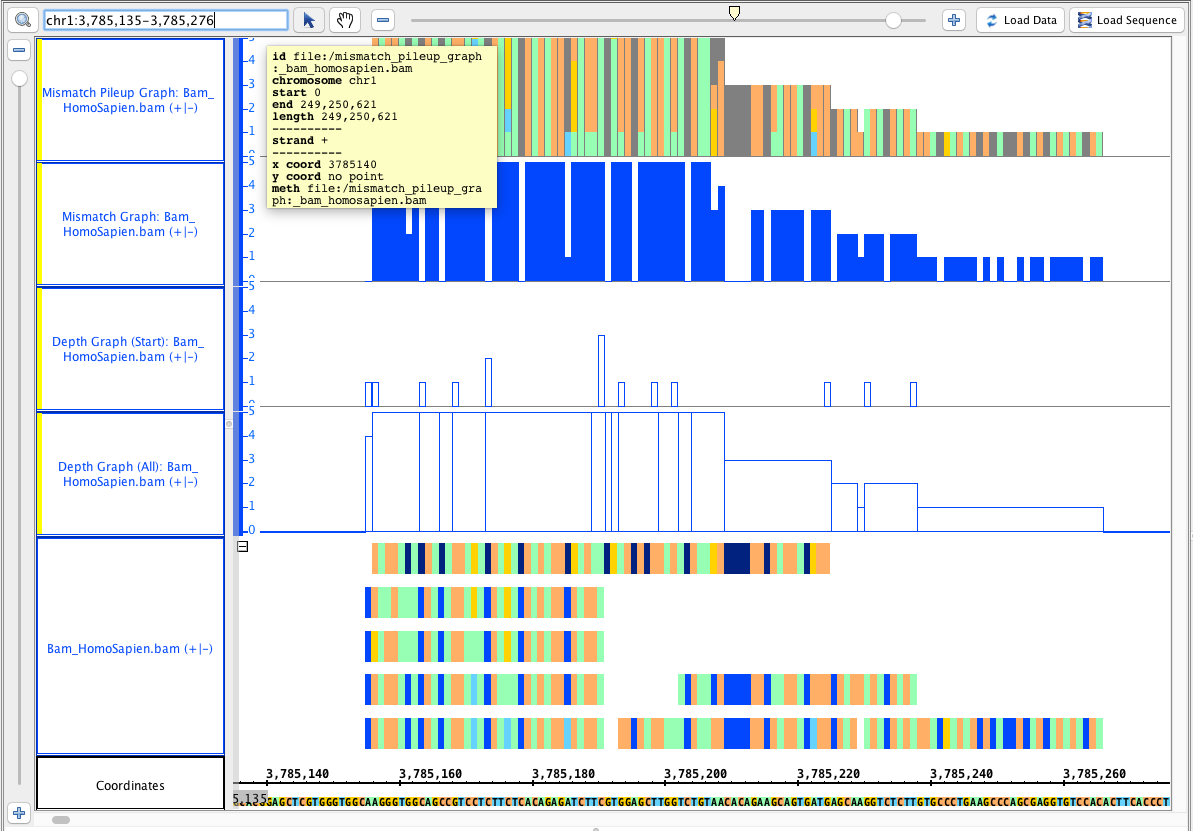
Users Guide







