General Function Checklist
- Download and unzip the following folder: test genomes.zip
- The folder "test genomes" should contain the following files:
- E_unicornis_Jul_2043.bed.gz.tbi
- E_unicornis_Jul_2043.bed.gz
- E_unicornis_Jul_2043.2bit
- T_rex_Jun_1993.2bit
- Start IGB 9.1.8 or later.
- Select the Plug-ins tab.
- Click Launch App Manager.
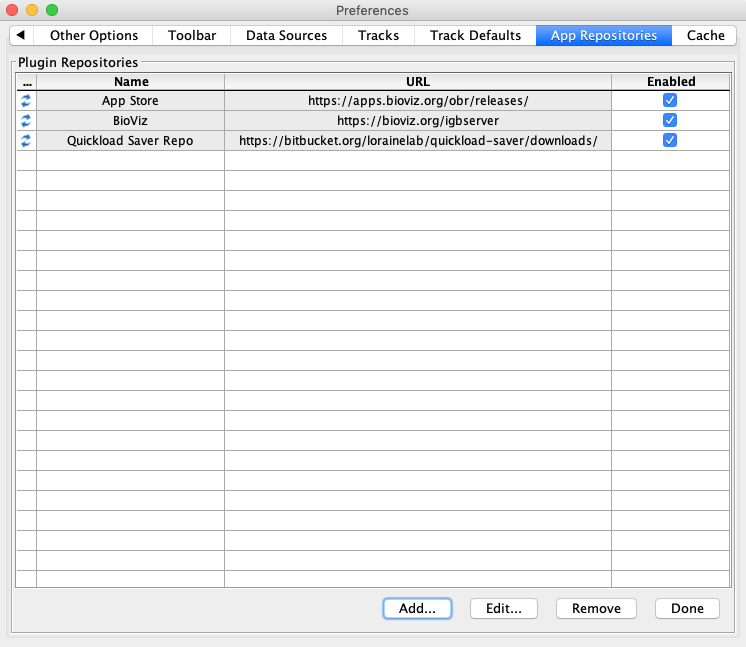
- Select Manage Repositories...
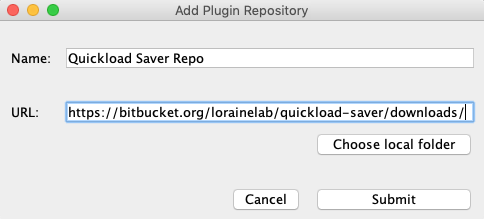
- Click Add...
- Name: Quickload Saver Repo
- URL: https://bitbucket.org/lorainelab/quickload-saver/downloads/
- Click Submit.
- Click Done.
- Checkpoint:
- mac
- linux
- windows
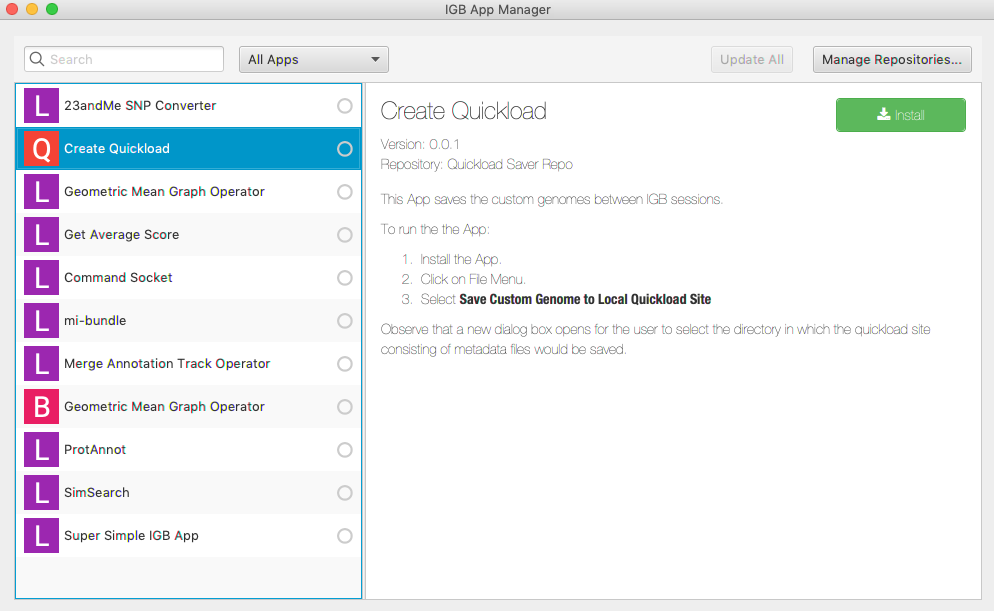
- In the Plug-ins tab, click Launch App Manager.
- Select the Quickload Saver app.
- Click Install.
- Checkpoint:
- mac
- linux
- windows
- Close the IGB App Manager window.
- Select File > Open Genome from File...
- Reference Sequence: Click Choose File... and select E_unicornis_Jul_2043.2bit
- Genus: Equus
- Species: unicornis
- Variety: (leave blank)
- Genome release date: July 2043
- Click OK.
- Select File > Open File...
- Select E_unicornis_Jul_2043.bed.gz
- Click Open.
- In the Data Access tab at the bottom of IGB,
- Change the Load Mode to Genome for E_unicornis_Jul_2043.bed.gz
- Change the color of the foreground (FG) to red.
- Checkpoint:
- mac
- linux
- windows
- Select File > Save Custom Genome to Local Quickload Site
- Select your desktop as the destination and unicorn as the name.
- Click Save.
- Select File > Open Genome from File...
- Reference Sequence: Click Choose File... and select the T_rex_Jun_1993.2bit
- Genus: Tyrannosaurus
- Species: rex
- Variety: (leave blank)
- Genome release date: June 1993
- Click OK.
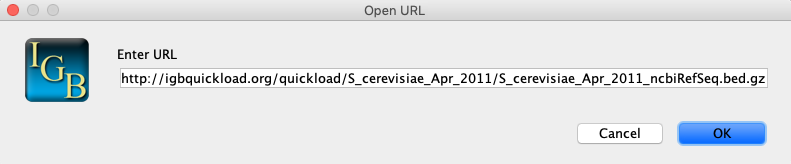
- Select File > Open URL...
- Enter URL: http://igbquickload.org/quickload/S_cerevisiae_Apr_2011/S_cerevisiae_Apr_2011_ncbiRefSeq.bed.gz
- Click OK.
- Checkpoint:
- mac
- linux
- windows
In the Data Access tab at the bottom of IGB:
- Change the Load Mode to Genome for S_cerevisiae_Apr_2011_ncbiRefSeq.bed.gz
- Change the color of the foreground (FG) to green.
- Checkpoint:
- mac
- linux
- windows
- Select File > Save Custom Genome to Local Quickload Site.
- Select your desktop as the destination and rex as the name.
- Select File > Preferences...
- In the Preferences window select the Other Options tab.
- Click Reset Preference to Defaults and then click Yes. IGB will close.
- Start IGB.
- Select File > Preferences.
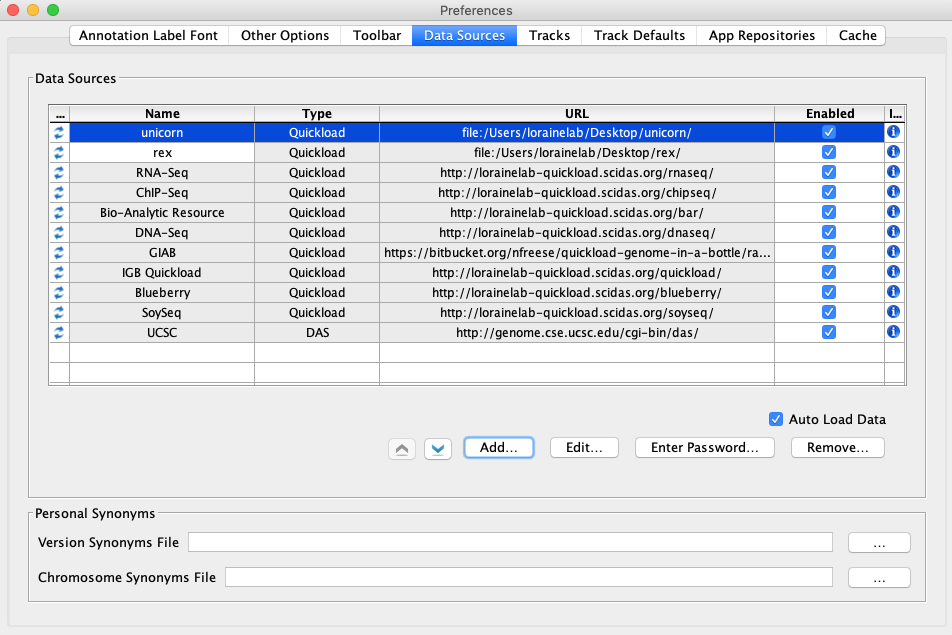
- Select the Data Sources tab.
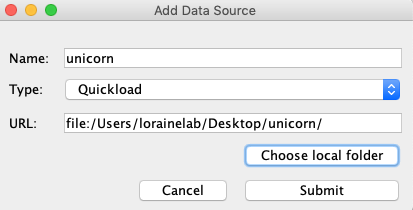
- Click Add...
- Name: unicorn
- Type: Quickload
- Click Choose local folder
- Select the unicorn folder on your desktop and click Open.
- Click Submit
- Checkpoint:
- mac
- linux
- windows
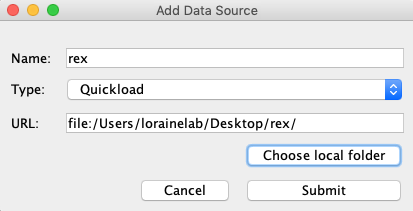
- Click Add...
- Name: rex
- Type: Quickload
- Click Choose local folder.
- Select the rex folder on your desktop and click Open.
- Click Submit.
- Checkpoint:
- mac
- linux
- windows
- Close the Preferences window.
- Select the Log tab and click Clear.
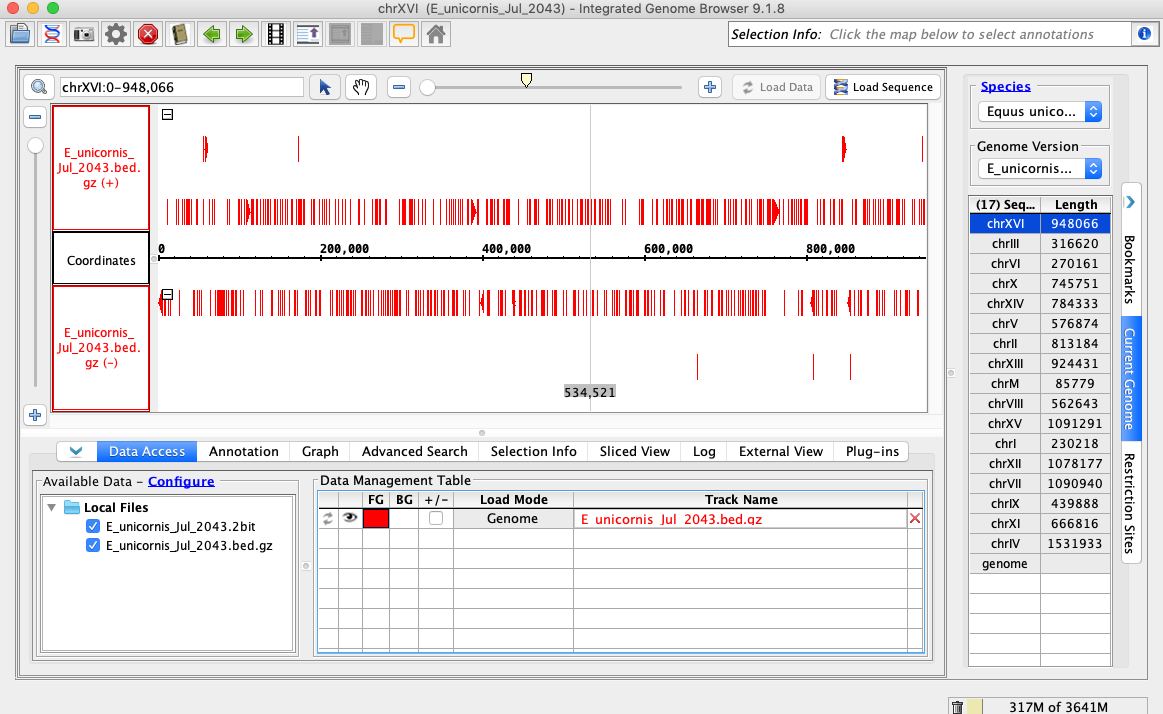
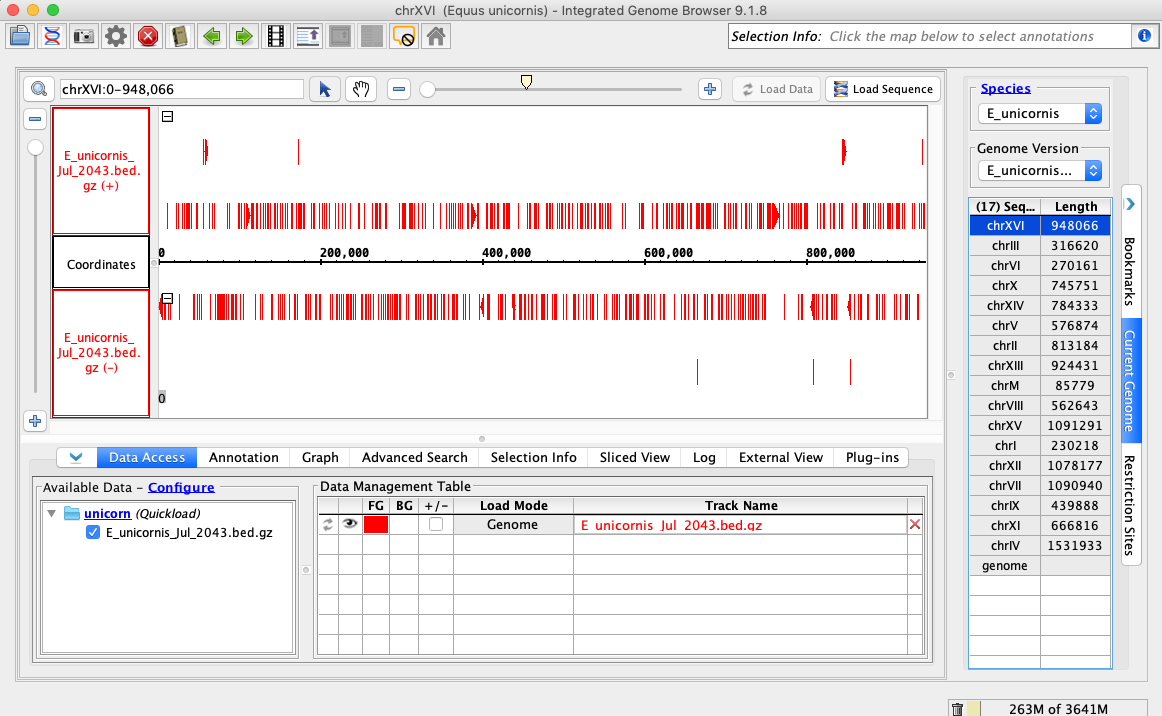
- Open the E_unicornis_Jul_2043 genome.
- In the Data Access tab, E_unicornis_Jul_2043.bed.gz should be set to Load Mode Genome and should be red (FG).
- Your IGB appears to look the same as the above image.
- mac
- linux
- windows
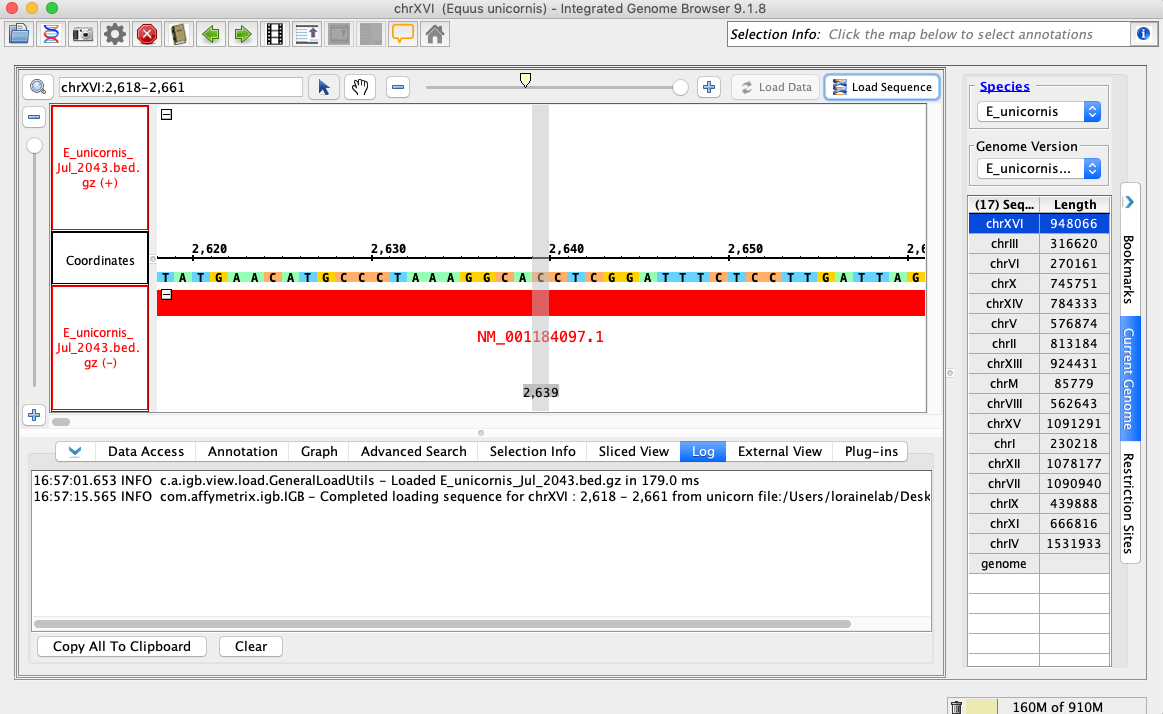
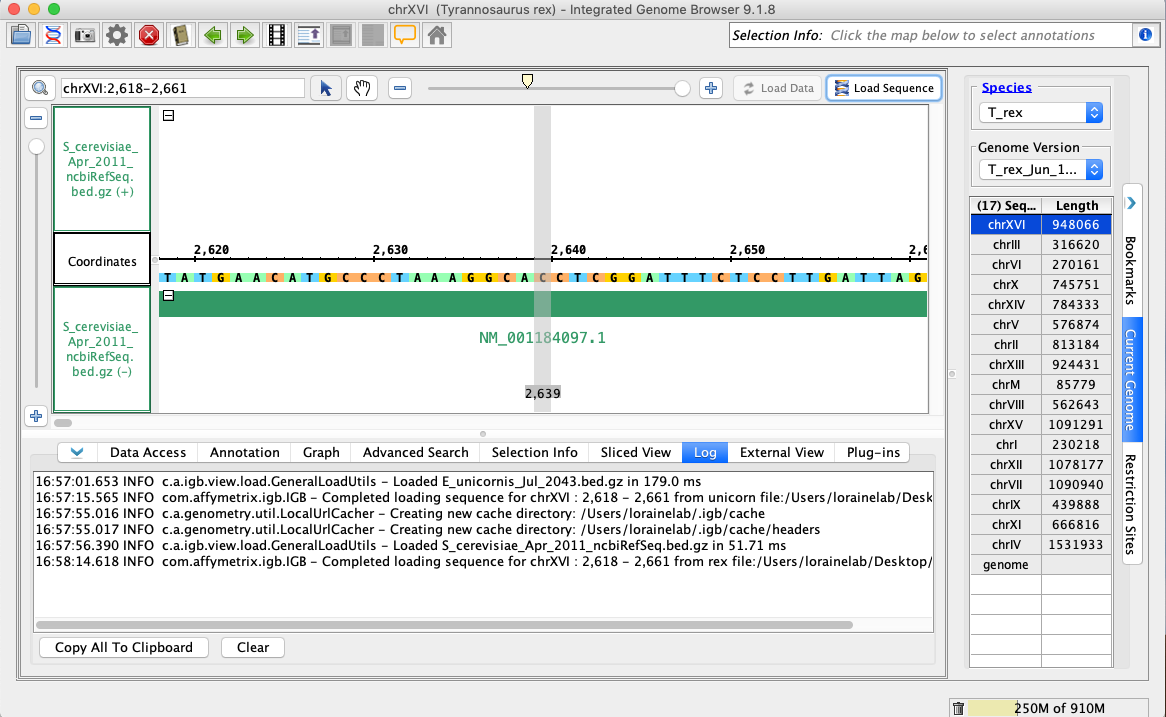
- Navigate to: chrXVI:2,618-2,661
- Click Load Sequence.
- Your IGB appears to look the same as the above image.
- mac
- linux
- windows
Select the Log tab and check for errors.
- There are no errors in the Log.
- mac
- linux
- windows
- Click Clear in the Log tab.
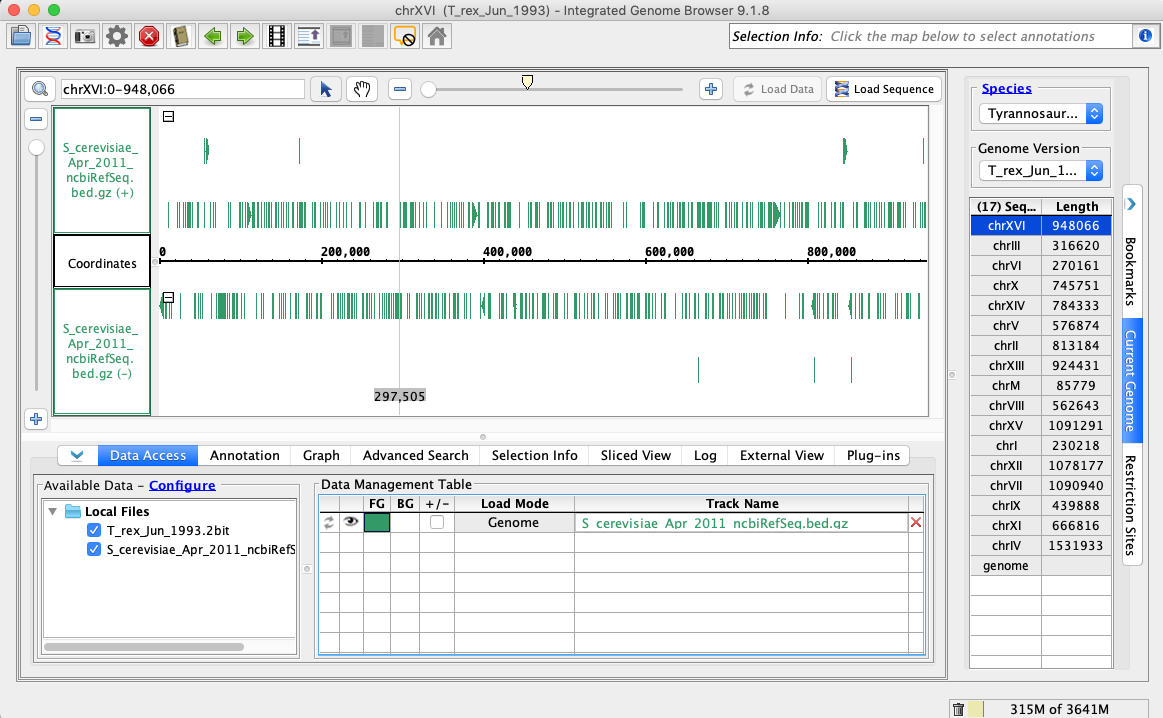
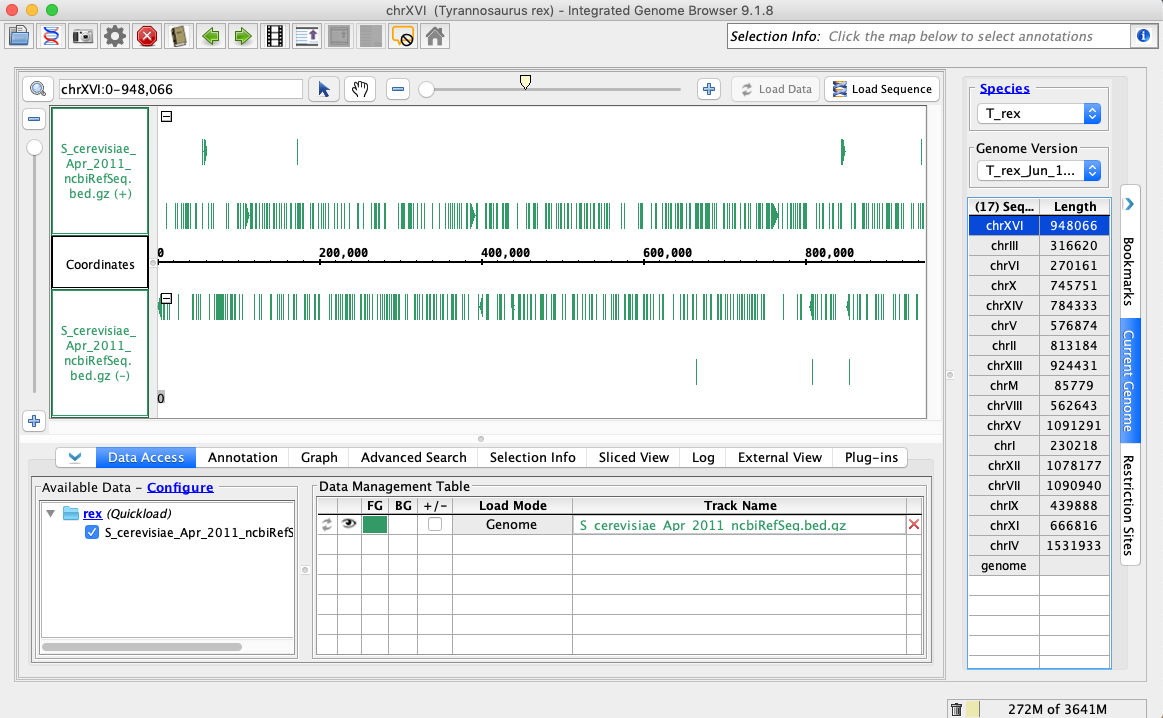
- Open the T_rex_Jun_1993 genome.
- In the Data Access tab, S_cerevisiae_Apr_2011_ncbiRefSeq.bed.gz should be set to Load Mode Genome and should be green (FG).
- Your IGB appears to look the same as the above image.
- mac
- linux
- windows
- Navigate to: chrXVI:2,618-2,661
- Click Load Sequence.
- Your IGB appears to look the same as the above image.
- mac
- linux
- windows
- Select the Log tab and check for errors.
- Click Clear in the Log tab.
- There are no errors in the Log.
- mac
- linux
- windows