Skip to end of metadata
Go to start of metadata
You are viewing an old version of this page. View the current version.
Compare with Current
View Page History
« Previous
Version 34
Next »
General Function Checklist
- ProtAnnot is launched from IGB
- Install the ProtAnnot plugin through the App Manager (Tools>Open App Manager)
- Select any genome
- Select one or more whole gene model annotation
- Please note, to simulate the images in this testing module, select gene model annotations that are adjacent to one another, rather than annotations that are far apart. Selecting annotations that are far apart will cause ProtAnnot to display a view that is significantly more zoomed out compared to the images in this testing module. However, this can be adjusted with the zoom bar feature.
- Select the Tools Menu
- Select Start ProtAnnot
- Checkpoint
- Your selected gene model(s) should look as follows when loaded into ProtAnnot; take note of the following key items in red:
- The window title
- The ProtAnnot menu
- The navigation and zoom bar
- Th selected gene model(s) name
- The Properties and InterProScan tabs
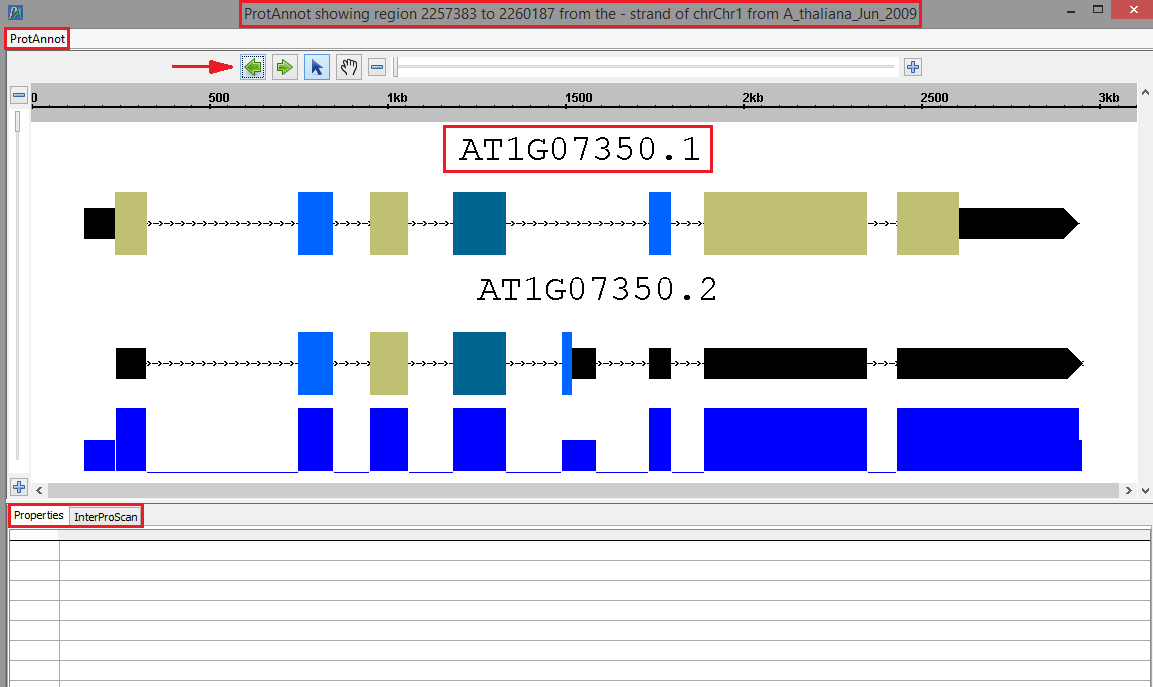
- The ProtAnnot window title should contain the absolute sequence coordinates, strand direction, contig name, and the genome version.

- InterProScan
- Select the ProtAnnot menu
- Select Run InterProScan menu item
- Verify that the InterProScan Job Configuration window appears similar to the image below
- Providing a valid email address is required for running InterProScan. Otherwise, InterProScan does not return any jobs.
- For testing purposes, please note that if an invalid email address is provided, the user may not see any jobs running.
- Verify correct text wrapping for the information icon
- Verify that the information link directs to the appropriate "About InterPro" page
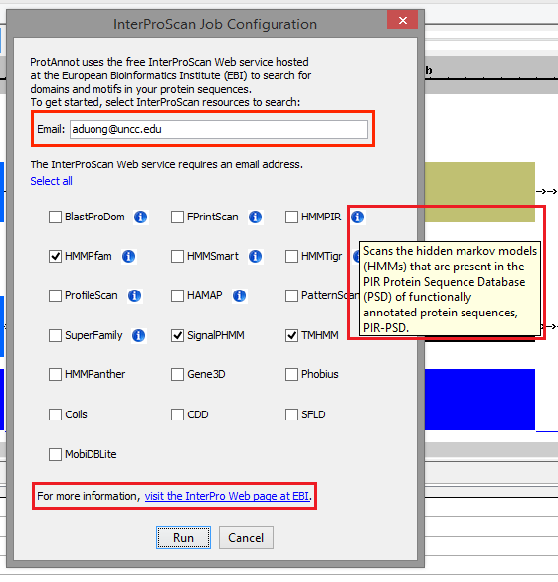
- Select Run
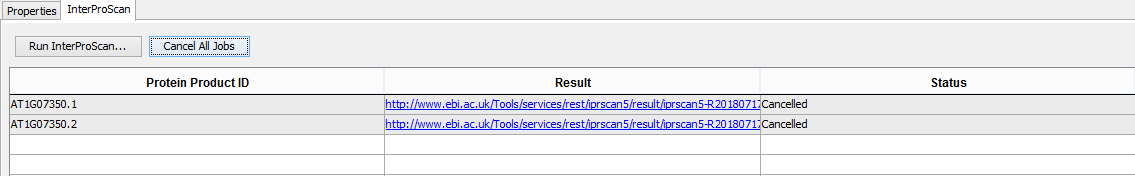
- Select the InterProScan tab
- Verify that all job statuses are completed with a status of FINISHED.
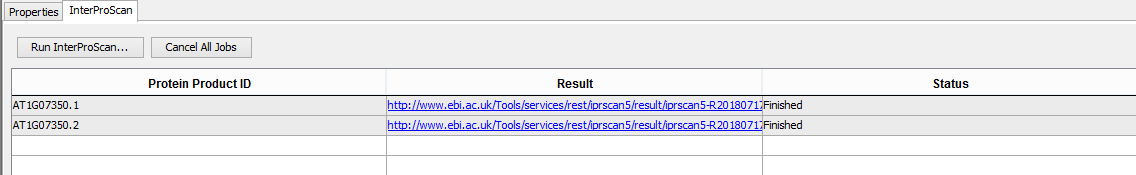
- Clicking on the result cell should open the result xml on the InterProScan website

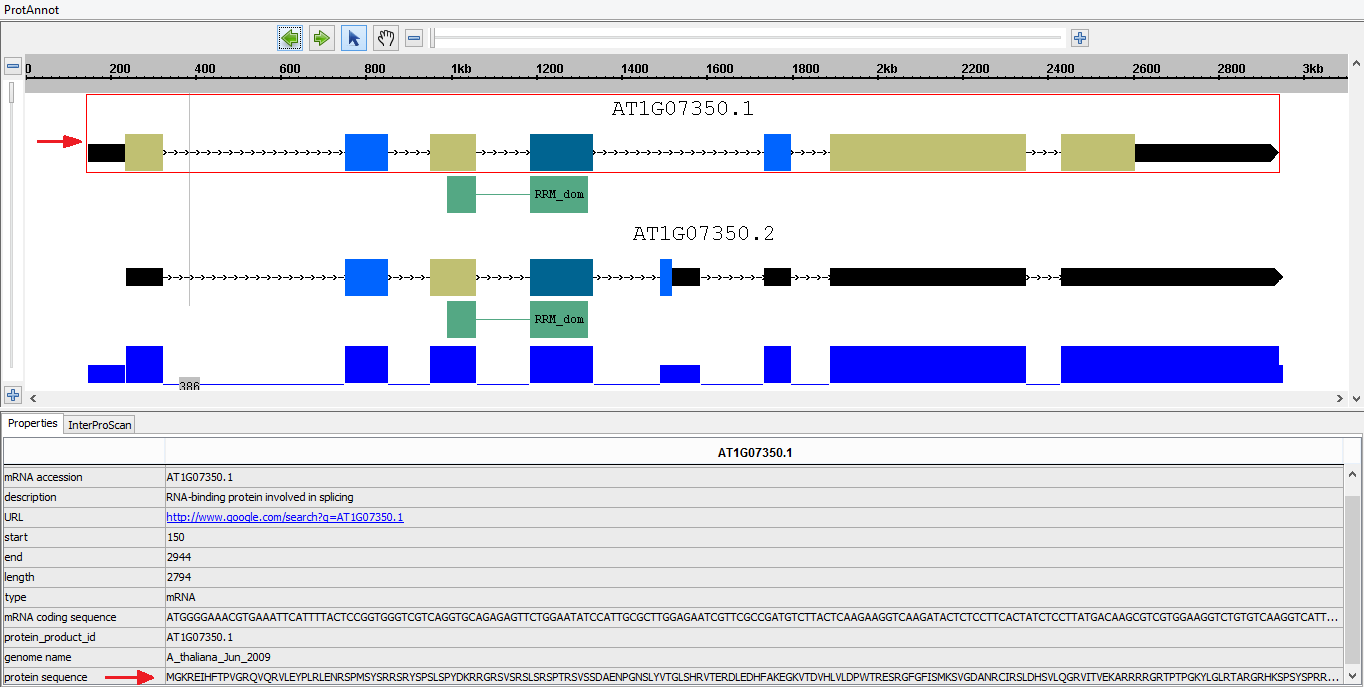
- Select the Properties tab
- In the display window, select a gene model to open the Properties tab, and verify that exons have the correct protein sequences.
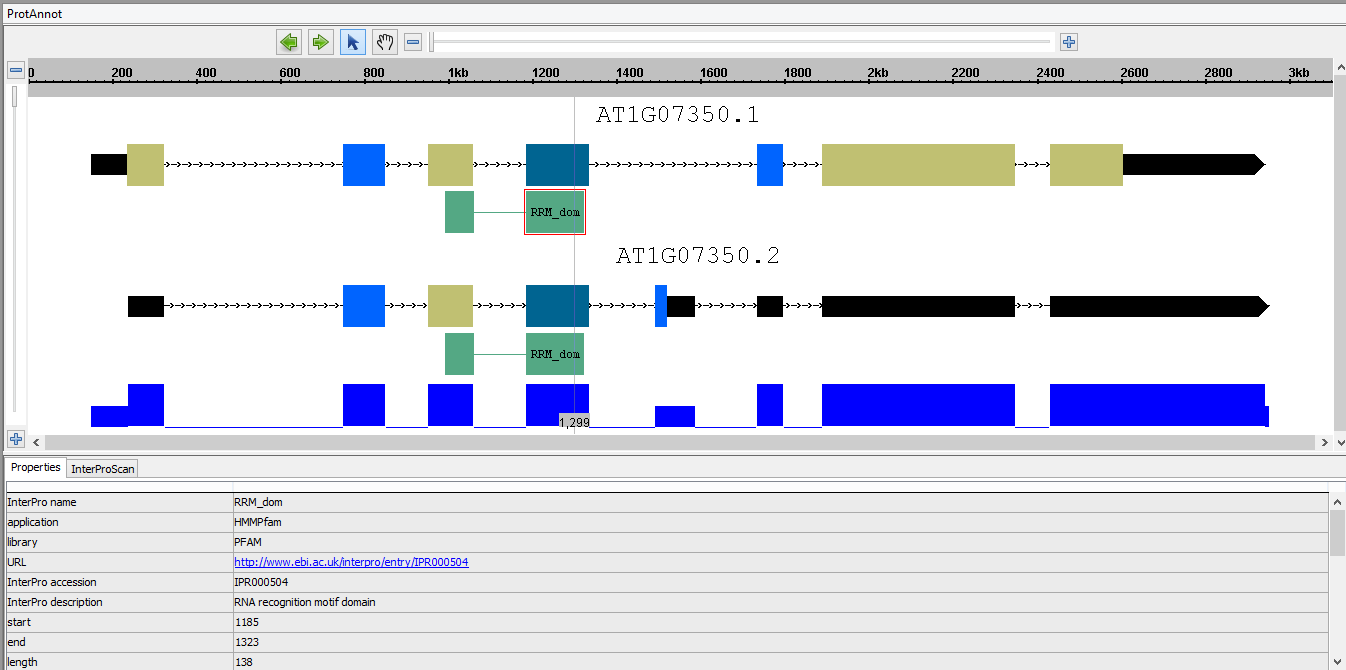
- In the display window, select a protein annotation to open the Properties tab, and verify that the following properties are present and correct:
- Application and library name
- Functional URL link
- Start, end, and length coordinates
- Run the InterProScan again by selecting Run InterProScan under the InterProScan tab
- Once the jobs are running, select Cancel All Jobs under the InterProScan tab
- Verify that job statuses are CANCELLED, and that there are no exceptions in the logs