Skip to end of metadata
Go to start of metadata
You are viewing an old version of this page. View the current version.
Compare with Current
View Page History
« Previous
Version 93
Next »
Complete the following general function checklist to verify that the Search subsystem is operating as expected.
- Select the Homo sapien genome and type "dmd" in the Quick-search box.
- Verify that "DMD" is one of the suggested matches in the Quick-search drop-down.
- Mac
- Linux
- Windows
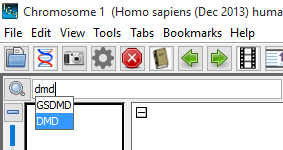
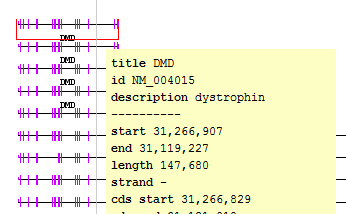
- Select "DMD" from the Quick-search drop-down to navigate to the longest known gene in the human genome, the dystrophin gene (DMD).
- Verify that IGB has navigated to the proper gene (dystrophin) by viewing the selection info for the gene. The gene's title should be "DMD".
- Mac
- Linux
- Windows
- Select the Advanced Search tab.
- Verify that search results for "DMD" are already being displayed in the tab.
- Mac
- Linux
- Windows
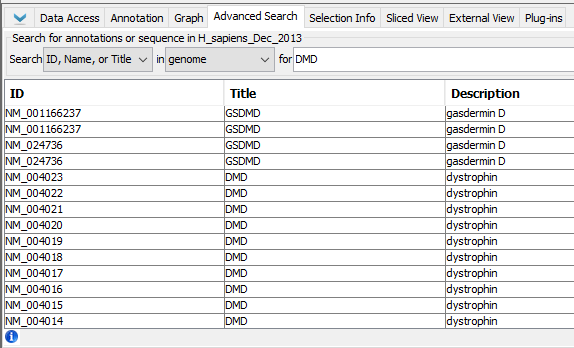
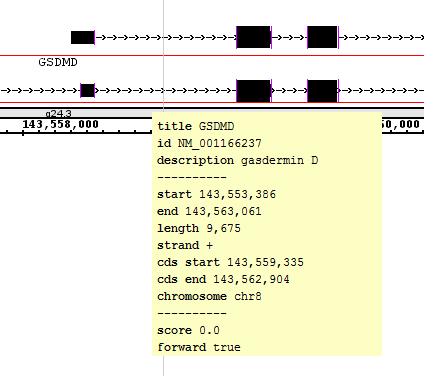
- Select the top search result in the Advanced Search tab (ID: "NM_001166237", Title: "GSDMD").
- Verify that IGB has navigated to "NM_001166237" in the gene "GSDMD".
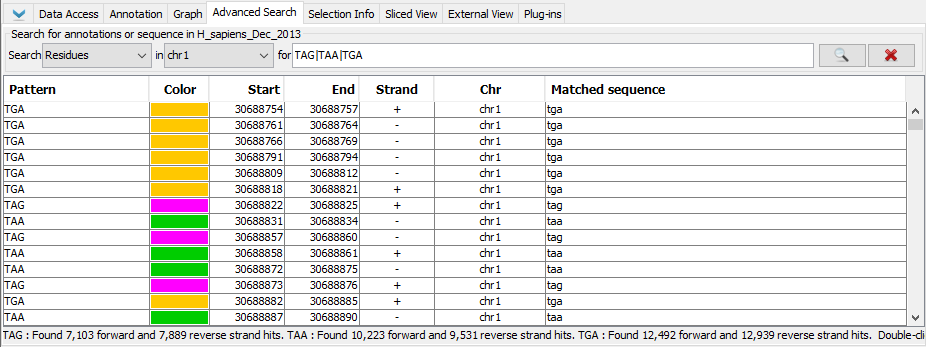
- Enter "chr1:30,688,740-31,361,080" into the Quick search box to navigate to the specified coordinates.
- Verify that IGB has navigated to the proper location.
- Mac
- Linux
- Windows
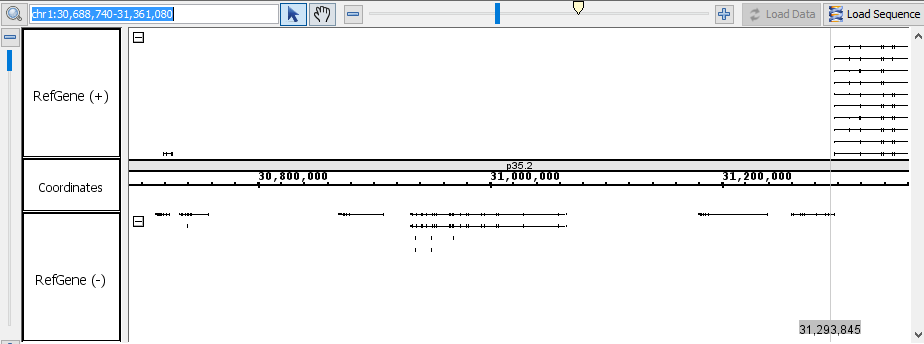
- In the Advanced Search tab, select Residues from the search type drop-down menu.
- Enter "TAG|TAA|TGA" into the Advanced Search textbox and start the search.
- Verify that the search has located matches for "TAG", "TAA", and "TGA".
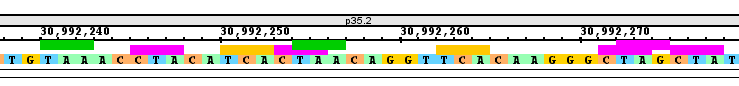
- Enter "chr1:30,992,236-30,992,279" into the Quick search box to navigate to the specified coordinates.
- Verify that residues have been highlighted on the sequence axis.
- Mac
- Linux
- Windows
- Test regular expressions. See the table in the users guide for explanations. Go to "chr1:30,688,740-31,361,080". In the Advanced search Panel, select Residues. Clear the search results between each search (the red X).
- Enter "TATATttatg"
- Enter "TATA.ttatg"
- Enter "TATA..tatg"
- Enter "TAT[AT]Tttatg"
- Enter "[TA]{1,3}Tttatg"
- Enter "TATAt*atg"
- Enter "TATAT.*ttatg"
- Enter "TATAT.*?ttatg"
- In the Advanced Search tab, select Properties from the search type drop-down menu.
- Enter "false" in the search text box.
- Verify that there are many results and if you select one (double-click it) and go to the Selection Info panel, there is at least one attribute (probably the called "forward") that has the value "false".
- Enter "family" in the search text box.
- Verify that there are many results and all include the word "family" in the description. (if not, check the selection info panel).
User Guide
- Verify that the relevant page in the Users Guide (Advanced Search) appears accurate and up-to-date.
The general function checklist for the Search subsystem has been completed.






