Introduction
IGB aims to be a truly integrated genome browser, meaning it can display data from a variety of diverse data sources, all merged into can integrate and merge data from multiple sources in the same view. This includes These include data sets loaded from your computer, from URL sites, or from various public (and private) DAS, DAS2, UCSC REST, and Quickload servers. IGB can also display data from many file types, including:
- Standard annotations such as RefSeq annotations, provided by public repositories.
- Alignments of Affymetrix probe sets to the genome, provided by the NetAffx group at Affymetrix.
- Alignments of 454 data.
- Tiling array graphs, from the TAS program.
- Copy number graphs from the CNAT program.
- Data generated from other Affymetrix software tools, such as GCOS, Expression Console and ExACT.
- Annotation and graph files prepared by any method in any of the supported formats.
The full list of supported file formats is here.
Choose Species and Genome Version
The first step to loading data is to choose Species and Genome Version. IGB uses this information to offer data sources with relevant data sets. To set the species and version, select them in the Data Access panel.
Alternatively, if you chose to open a file from
To load data:
| Table of Contents |
|---|
...
Choose Species and Genome Version
Use the Current Genome tab to select a Species and Genome Version, or select an image on the Start Screen.
You can also open custom genomes not already available in IGB. See Custom Genomes (Genomes not in IGB).
...
Open data sets - from files, URLs, Quickload sites, UCSC Genome Browser, or other sources
Once you select a species and genome version, publicly accessible data sets hosted by UCSC Genome Browser, IGB Quickload sites, and other sources will appear in the Available Data section of the Data Access tab. Select these data sets to open them in IGB.
You can also open files in IGB if they're stored locally or via a publicly-available URL. To open a file, select File > Open File... or File > Open URL... the file selection widow will permit you to specify species and genome using the drop down menus at the bottom of the window.
While some file types contain information specifying their species and genome version, most do not. However, if you load a file that has the species and genome information, IGB will open the proper species and genome, even if it is not the one you have chosen.
Loading Data Sets
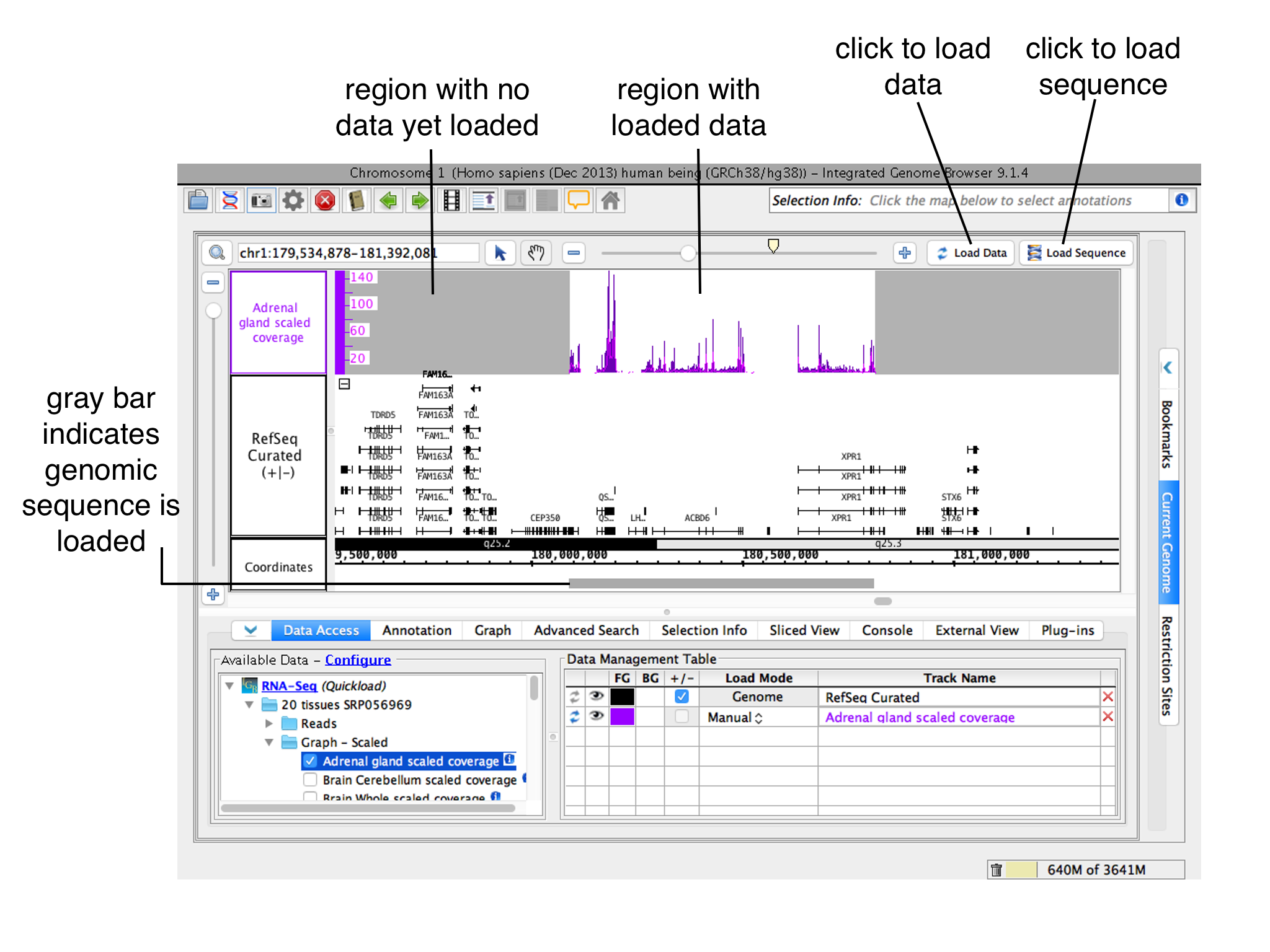
NOTE: IGB does NOT immediately display loaded files. Many of today's next-gen sequencing files are too big to display all at once. IGB handles this issue by waiting to visualize data sets until you ask it to refresh. While you can immediately refresh to visualize most files, many larger file types, such as BAM and WIG should first have a defined, smaller region selected prior to refreshing the image.
There are several ways to get data sets into IGB, from servers/sources, from URLs and from the local computer. To load data from a server, locate the data set in the folders of the Data Sources panel. Put a check in the box next to the data you are interested in. This file will be entered into the Choose Load Mode list.
For files loaded from URL or from the local computer, just drag and drop into the IGB interface; the file will immediately appear in the Choose Load Mode list. Alternatively, use the File > Open File.. or Open URL... to find and load the file(s) you want. Be sure to set the species and genome at the bottom of the file selection window..
Once a file is opened, IGB adds it to the Data Management Table in the Data Access tab.
| Note |
|---|
Opening a file does not automatically cause IGB to start showing data in the file. Because some data files are very large, you must first zoom in and click the Load Data button load the data into IGB. |
...
Use Data Management Table to configure data loading settings (optional)
To change how and when IGB loads data into a track, open the Data Access panel and change settings under Load Mode in the Data Management Table.
Options include:
- Don't Load means no data will load when you click the Load Data button.
- Genome triggers automatic loading of all data from a file.
- Manual means that only data overlapping for the currently displayed region will load when you click Load Data. This is the default setting.
- Auto means that moving or zooming to a new location triggers data loading, but only when the zoom threshold is lower than the mark shown on the horizontal zoomer. Change the threshold by selecting View > Set AutoLoad Threshold to Current View.
...
Click Load Data or Load Sequence buttons
To load data for all visible tracks with load mode setting Manual, click the Load Data button.
To load reference sequence, click the Load Sequence button.
To load all data for a data set, change its Load Mode in the Data Management Table to Genome.
Regions where data have not yet been loaded have gray background color.