| Table of Contents |
|---|
...
Probe set alignments - link.psl files
IGB can be used to visualize data from Affymetrix showing the locations of GeneChip Expression Array probe sets and target sequences aligned to a genome.
In most Affymetrix arrays, probes are grouped conceptually into probe sets, groups of probes that are expected to measure expression for individual known or computationally-deduced mRNA molecules.
These target sequences may be identical to known mRNA sequences in GenBank, or they may have been produced computationally by merging ESTs or mRNA sequences into a single sequence, sometimes called a "consensus" sequence.
IGB can display location of design sequences and probes within the genomic sequence. Probe set alignments are available as "link.psl" files from the Affymetrix Web site. Some probe set alignments are also available from Affymetrix and IGBQuickLoad data sources in the Available Data sub-panel of the Data Access planel.
Viewing probe sets from IGB DAS
To view probe sets available from IGB DAS2 data source
- Open the Data Access panel
- Open the IGB DAS / affy folder under the Data Sources and Data Sets section of the Data Access tab.
- Click the checkbox next to the array you would like to examine
- zoom to a gene or region of interest and click Load Data.
The probe set, including the target sequence and individual probe, will appear in IGB.
Where to find probe set alignment files
Many are available from the Affymetrix customer support Web site.
To obtain these files, go to the Affymetrx site and look for the array you are interested in.
On the array's support page, look for alignment data files in the "NetAffx Annotation Files" section. Look for files labeled as consensus alignment files or something similar. Note that alignment data files may not be available be available for species with well-characterized genomes.
Affymetrix distributes these files in an Affymetrix-specific format called "link.psl" which IGB can read.
These files consist of two sections:
- Alignments of the target sequences onto the reference genome (ordinary PSL format)
- Alignments of probe sequence onto the targets
Probes and probe sets on display
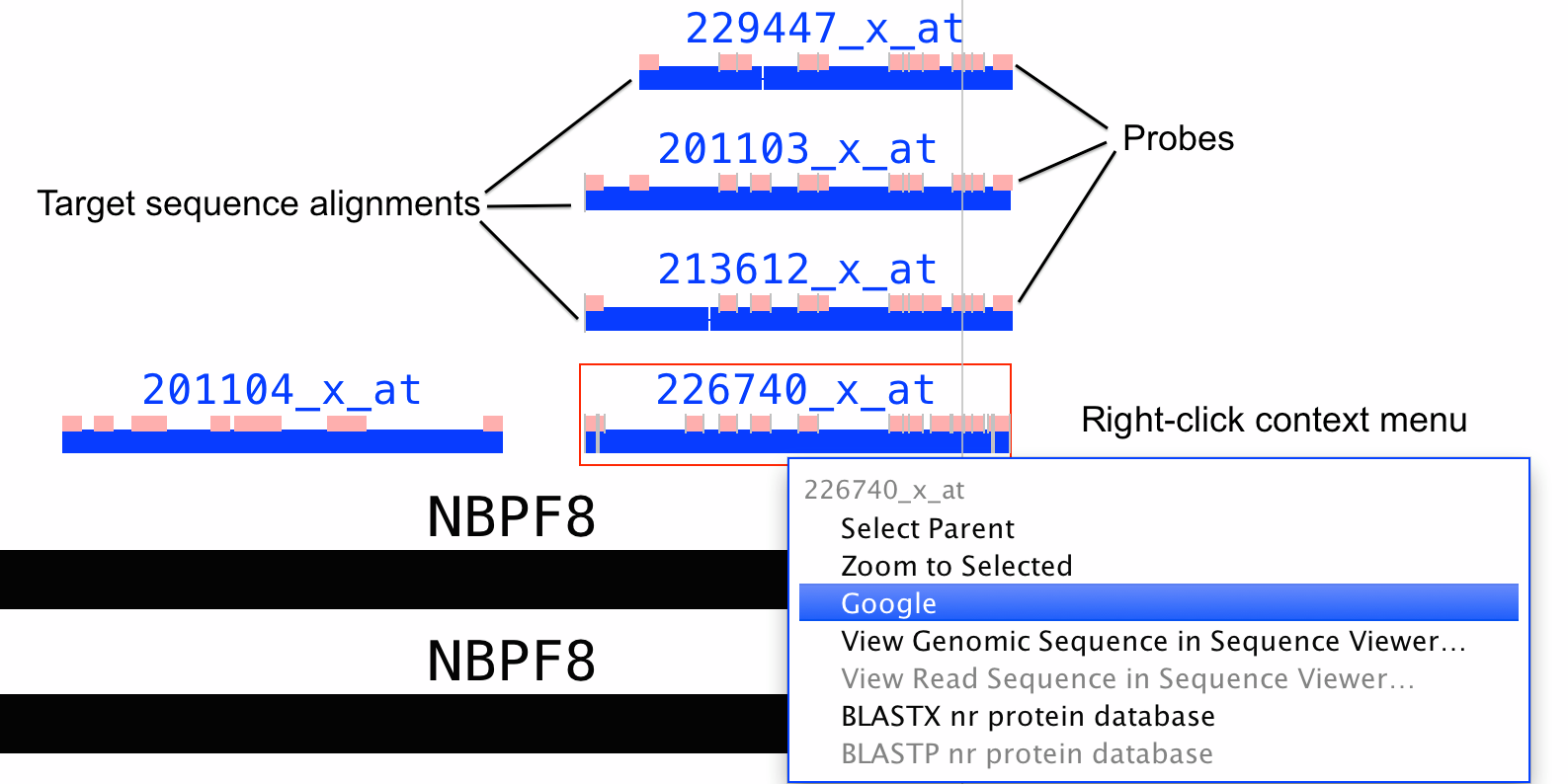
Once the data have been loaded, the probes and target sequence alignments will appear in a separate track labeled by chip. Within the track, probes and their matching target sequences always appear together. The following figure shows an example from the rat 230 chip.
Probe set target sequence
The alignment between the probe set target sequence and the genome is represented at the top of the figure as a series of blocks. Each block represents a block of alignment in which each base in the genome matches a corresponding base in the target sequence. Gaps between the blocks typically represent areas where the genomic sequence contains inserts relative to the aligned target sequence. Usually, these gaps are due to introns.
There are some exceptions to this, however. For example, the sixth and seventh blocks in the figure above are so close together that they almost appear as a single block at this level of zoom.
Zooming in for a closer view reveals that these two alignment blocks are immediately adjacent to each other. This indicates that these two blocks of alignment were separated by an insert in the target sequence relative to the genomic sequence. That is, the target sequence contained some bases that were not present in the genomic sequence. This may present a problem if this missing region (in the genome, that is) contains some probe sequences. In this particular case, however, the alignment irregularity occurs in a 5' region, outside the area covered by the probes. (See the discussion below.)
How can this happen? There are a number of reasons, but discrepencies between the genomic and target sequences are usually responsible.
Probes and probe sets.
Each target sequence is shown with its corresponding probe set. Each probe set consists of a group of probes, which are shown superimposed on the alignment blocks of the target sequence.
The figure below shows a close-up view near the 3' end of the target sequence. The 3' end of the target sequence is annotated with blocks which represent individual probes.
Two things are important to notice about this image. First, sometimes individual probes are split across gaps in the alignment, which typically correspond to introns. When this occurs, the two halves of the probe are connected by a line.
Second, sometimes probes overlap with each other. This can be seen by clicking on probes. If a selection outline is visible in the middle of what otherwise looks like a single block, then there is really more than one block.
Probe set labels
Each probe set and probe set target sequence is labeled with the name of the chip (Rat230_2, in this case) and the probe set identifier.
Polyadenylation sites
For many probe set target sequences, Affymetrix bioinformatics scientists have used computational methods to predict putative polyadenylation sites near the 3' end of the target sequence. These are shown as dark or light blue boxes riding on top of the terminal alignment span.
If the box is light blue, then the site was deduced from the overlap of multiple expressed sequences (usually 3' ESTs) whose genome alignments all terminate at a common location. This is sometimes called a "polyA stack."
If the box is dark blue, then the site has been deduced through sequence analysis of an individual, exemplar sequence, such an mRNA sequence record from GenBank. This is sometimes called a polyA site to distinguish it from a polyA stack.
Recognizing putative probe set targets
It is usually a good idea to load additional tracks of data besides just the probe set information. For example, in the figure above, the track below the probe set track shows the probe set's likely target, an mRNA from the Rp14 locus. Based on their relative alignments to the genome sequence, it appears that both sequences overlap in the region that contains the probes. Thus, it is very reasonable to assume that this probe set does indeed detect the mRNA shown below it.display Affymetrix probe sets aligned onto a reference genome - it can show probe set design sequences aligned onto a genome with the locations of the probes indicated as blocks.
When IGB was first developed at Affymetrix, the company distributed probe set alignment files for its catalog 3' IVT arrays. In recent years, however, they've stopped updating these files. So for some genomes, the alignment files you can find on the Affymetrix Web site reference obsolete reference genomes. If you need to work with more up-to-date genomes, we recommend you create your own alignment files or request them from the IGB team. For some genomes, we've added probe set alignments to the main IGBQuickLoad.org site. Mainly we've done this at the request of individual researchers, and so if you would like to request an array, let us know. If probe set alignments are available from our site, you'll typically find them in a folder named Affymetrix under the Data Sources section of the Data Access panel.
You can also make your own probe set alignment files using blat, tabix, and a python script we wrote. For more information, see this Bitbucket repository.
About probe set alignment visualizations
Depending on when the arrays were designed, Affymetrix typically used expressed sequences from GenBank to select probes for probe sets - these expressed sequences were sometimes called "exemplar" or "consensus" sequences. They then selected individual probes from regions near the 3' end of the expressed sequence. Affymetrix (as of 2014) distributes probe and target sequences on their Web site, where "target sequences" contain the 3' end regions from which the probes were selected.
Probe set visualizations in IGB show the alignments of target, exemplar, or consensus sequences onto the genome. They also show the locations of probes that were selected from the design sequence. See the preceding figure for an example.
Because probes were selected from the expressed sequences, sometimes a probe will be shown as split across an intron. Also, sometimes probes overlap. And sometimes probes may be missing. If the target sequence contains a region that can't align onto the reference, and if this unaligned sequence contains a probe, then that probe will not be shown.
If you have questions about what you see in a probe set alignment, let us know.
Why this is useful
Often multiple, seemingly redundant probe sets interrogate one gene. This situation mainly arises when a gene has multiple, alternative three-prime ends due to alternative splicing or alternative termination sites. If an experiment identifies genes where redundant probe sets are differentially expressed with different fold-changes or in opposite directions, this can indicate that the treatment affects splicing as well as the overall abundance of RNAs arising from the gene.
Thus if you observe redundant probe sets that give different or contradictory results, it's a good idea to view them in IGB and compare their alignment to annotated genes and transcripts.