Introduction
As you learn to use IGB, you'll find it offers many features that make it one of the best tools available for visualization and exploration of genomic data sets.
If you are new to IGB, the following six step use this Quick Start Guide will help you get started using IGB.
| Table of Contents |
|---|
Step 1:
...
Get and start IGB
...
- Go to
...
...
- and click
...
- Go to IGB Download Page
2. Click the IGB image to download and launch IGB. If your computer has enough memory, we recommend choosing the high memory (2 or 5 GB) option.
IGB Download - Small, medium, and large memory options
Step 2: Choose your species and genome version, if available
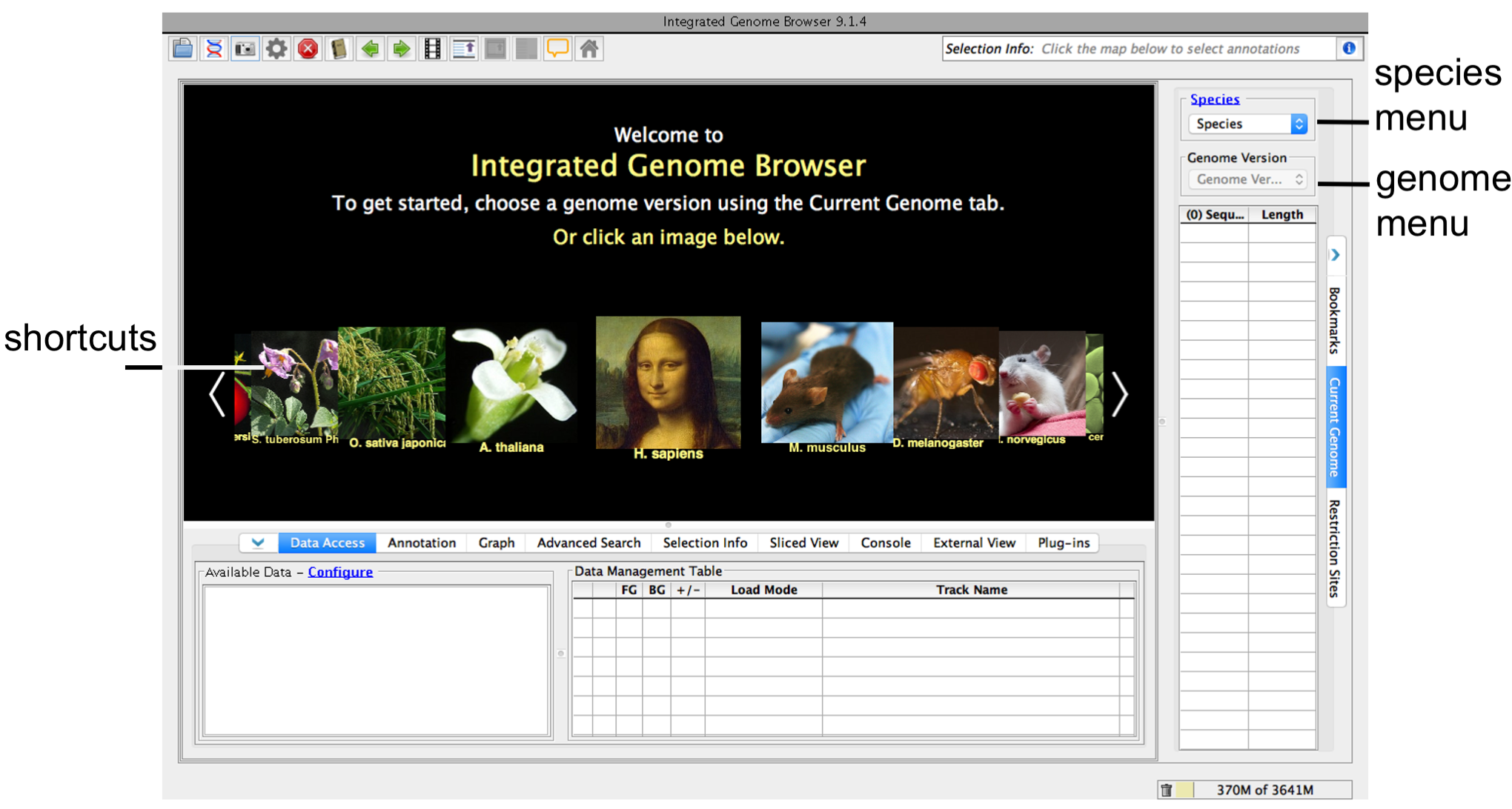
When IGB first starts, you'll see the IGB start screen shown below.
- Click an image or choose species and version under the Current Genome tab (blue box).
- For some species, gene models and cytological bands will automatically load.
- If your genome of interest is not listed, don't choose anything and skip to Step 3.
IGB start screen
IGB after selecting the latest human genome. RefSeq alignments are show in blue.
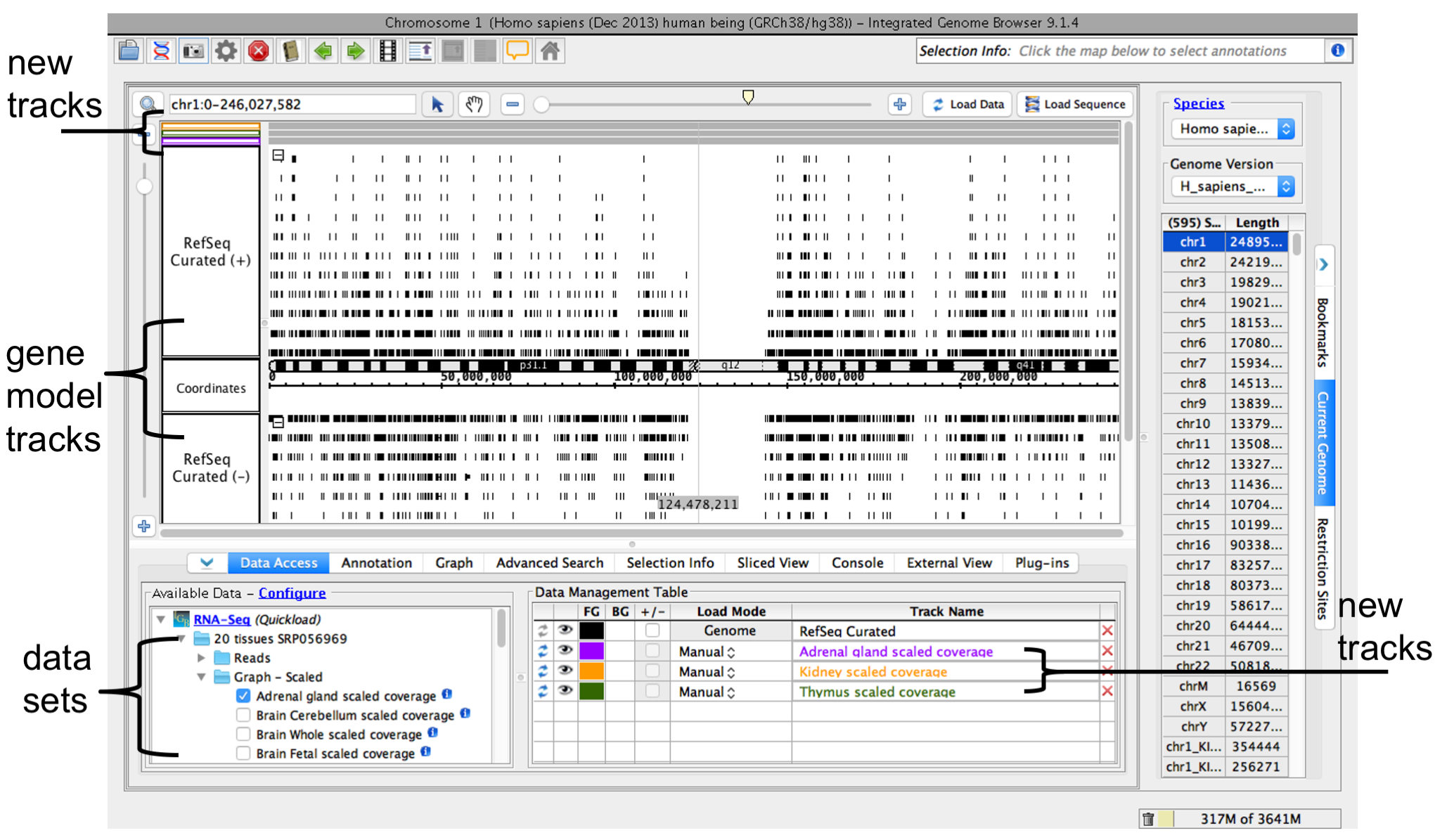
Step 3. Load data sets from Data Source servers
- Select and load annotations
- Open data source folders under the Data Sources and Data Sets file tree (red box)
- Select data sets you would like to view by
- checking the box
- or click-dragging them into the Data Management table
When you select a data the data sets you would like to load, IGB will add new track (orange box) to the main viewer.
To view data
- Zoom or scroll to a region of interest
- Click Load Data (green arrows, boxes) to load data into IGB
Step 4: Load data from local files
To view data from local or remote files
- Use File > Openthe Download button
- Select and download the installer for your platform
If you have trouble starting IGB, visit the help page on BioViz.org for assistance.
Step 2: Choose species and genome version
To choose a species and genome version
- Click a shortcut image (loads most recent genome)
or
- Choose Species and Genome Version using the Current Genome tab.
IGB start screen
If your species or genome version is not listed, you can view it if you have a fasta or 2bit file with sequence data. See Custom Genomes
Step 3. Open data sets
Open data sets from remote data sources (Data Access tab) or by opening local files.
To open a data set from an IGB Quickload data source (see About IGB Quickload)
- Select Data Access
- Select data sets in the Available Data section
To open local files on your computer
- Select File > Open File... or File > Open URL...
...
- Enter file name or URL
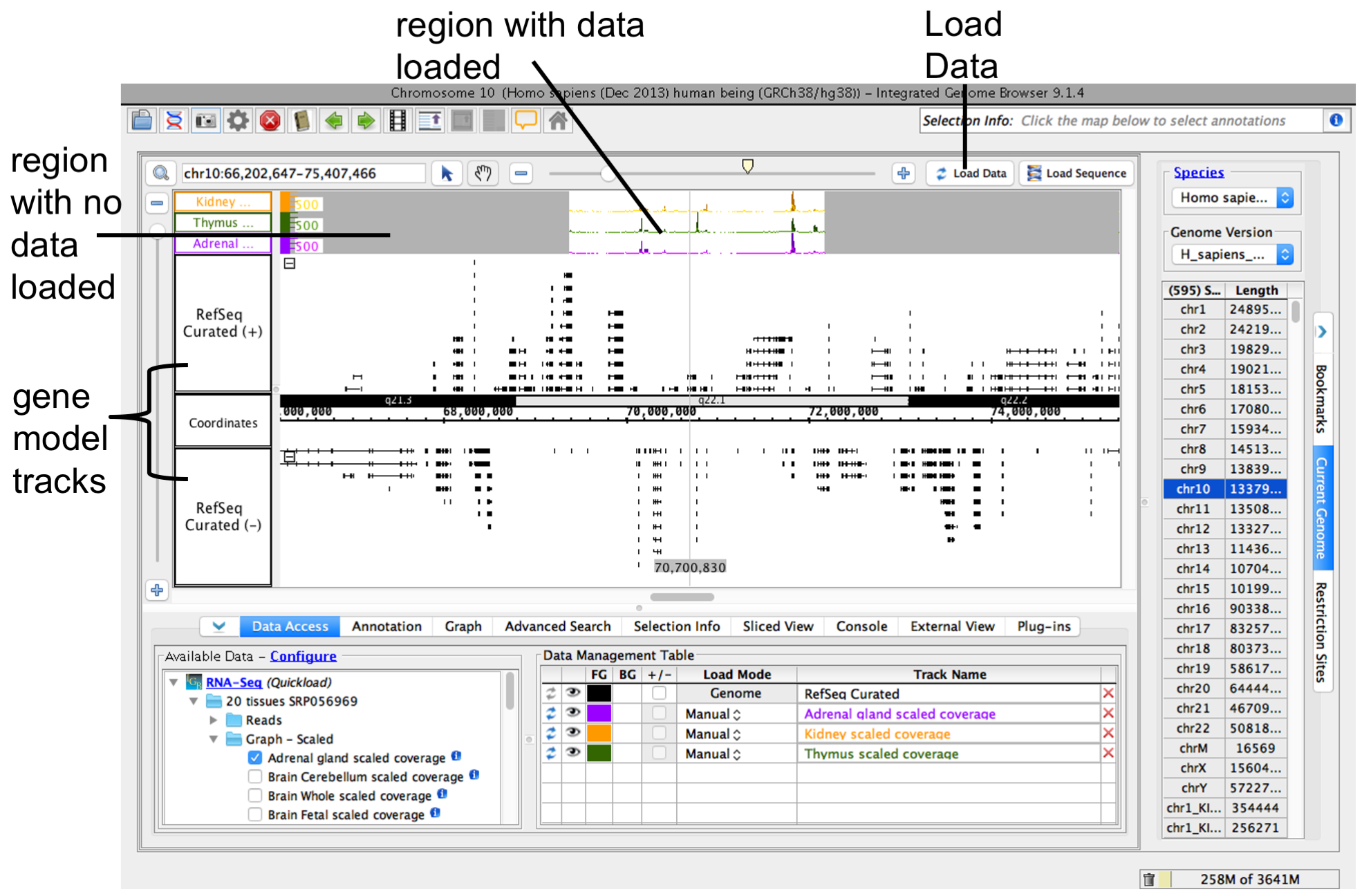
When you open select a data sets or a file, IGB adds a new empty track to the main view .
Areas that are NOT loaded are grayed out (as you can see in the place holder tracks).
and adds the file to the list of active data sets in the Data Management Table.
To load data into the main view, click Load Data.
Note: Many data files (especially BAM files) are huge and and lists it in the Data Management table. Empty regions in the new track that do not have data loaded are gray.
IGB window after opening human genome RNA-Seq coverage graphs from Adrenal Gland, Kidney, and Thymus data sets
Step 4: Zoom in
Because many data files contain too much data to display view all at once. For this reason, IGB does not automatically load data from a newly opened file. To view data in a file, zoom and scroll to a region or gene of interest and click Load Data. IGB indicates regions that have not yet been loaded by displaying a slightly darker background color.
Reference Sequence
If you have a sequence file (fasta, 2bit, bnib, etc.) that you would like to use as the reference sequence, use the File > Open Reference Sequence... option.
Step 5: Zoom in on a region of interest
If you're working with large files such as .bam, .sam, .wig, .bedGraph, you should 'zoom in' to a smaller area before loading your data, i.e. select a smaller region of data to load. To zoom in on a region of interest, you can:
- Use the zoom slider to zoom in on a gene (purple box/arrow). Click in the main window to focus zooming and then drag the slider to right to zoom in.
- Click the Search tab and enter the name of a gene. Double-click rows in the results list to jump to a feature (red box/arrow).
- See also Searching in IGB
- Double click on a feature within the visual field to zoom to that feature (orange box/arrow).
- See alsoSelecting a feature
- Enter a coordinate range in the coordinate box and hit enter to zoom to that location (green box/arrow)
- Click and drag within the coordinate axis to zoom to a region of interest (blue box/arrow).
- Use control+mouse wheel (Windows) or command+mouse wheel (Mac) to zoom in on the zoom stripe location (yellow box/arrow).
See also:
- More about scrolling/panning in IGB
- More about the main view
- More about the tabbed panels and about customizing them.
load data into the viewer until you click the Load Data button.
Before loading data, zoom in to a region.
- Click a location in the main view
- Drag the horizontal zoom slider or use plus and minus buttons
IGB horizontal zooming controls
Other ways to zoom
Other ways to zoom include
- Search for a gene by name or keyword (For example, TBATA or thymus)
- Double-click an exon or gene model to zoom in on it
- Click-drag the coordinate axis to zoom in on a region
See also:
Step 5: Load data
To load data, click Load Data button. Regions with loaded data show the selected background color; areas without loaded data appear darker.
IGB after loading data
See also:
Step 6: Configure tracks
You can reorder the tracks by dragging the Track Label (the Data Management Table reflect changes).
To change style elements of a track color, how IGB indicates strand, track labels, and much more.
To change track appearance
...
, click the track label and use the Annotation or Graph tab to change to change color, track height, annotation label, amount of data shown (stack height), and other options.
See also: