...
Step 1: Get and start IGB
- Go to BioViz.org and click the IGB Download Page button
- Select and download the installer for your platform
If you have trouble starting IGB, see Troubleshooting or ask for help on BioStars.visit the help page on BioViz.org for assistance.
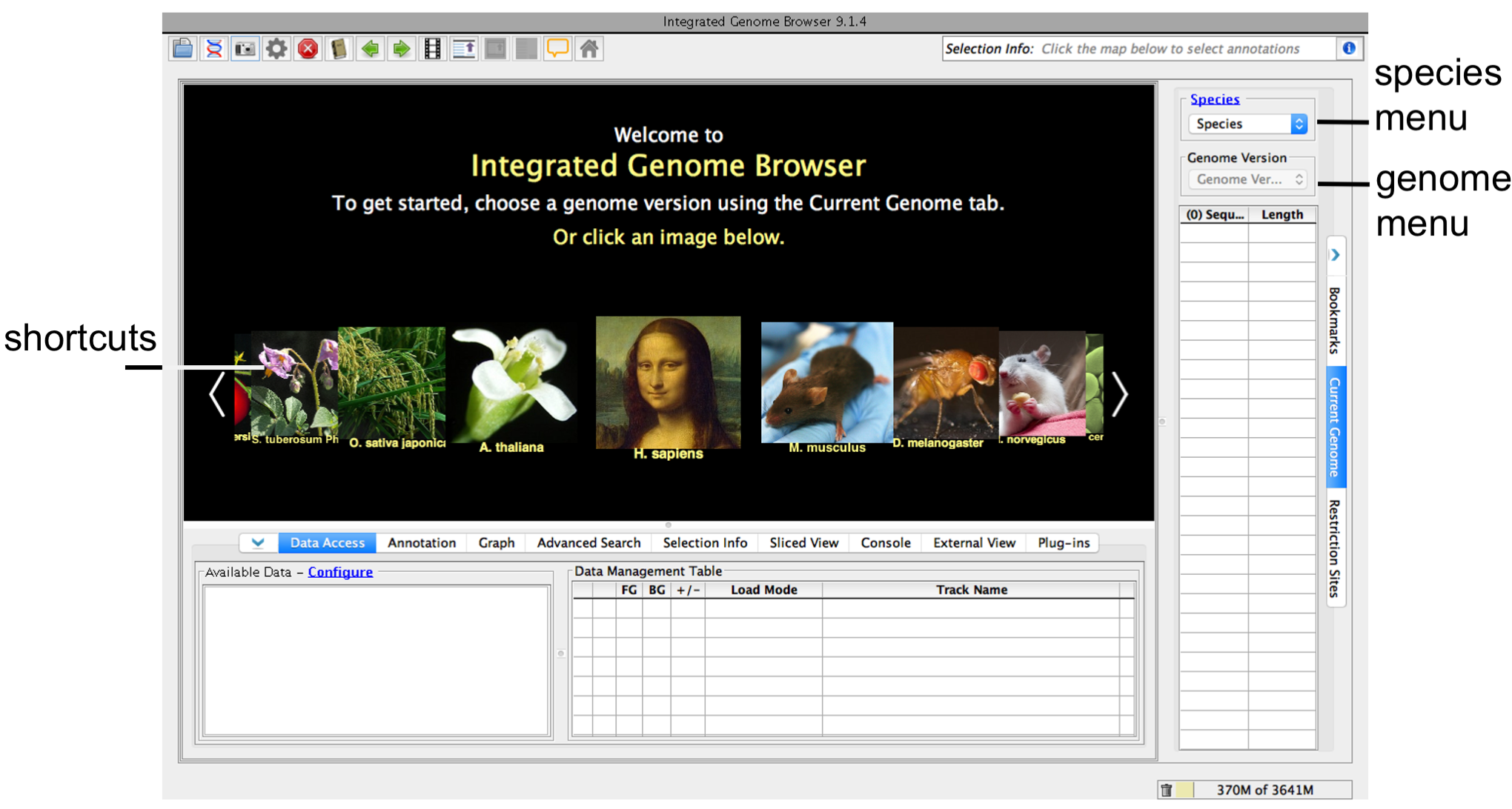
Step 2: Choose species and genome version
...
- Choose Species and Genome Version using the Current Genome tab.
IGB start screen
If your species or genome version is not listed, you can view it if you have a fasta or 2bit file with sequence data. See Custom Genomes
...
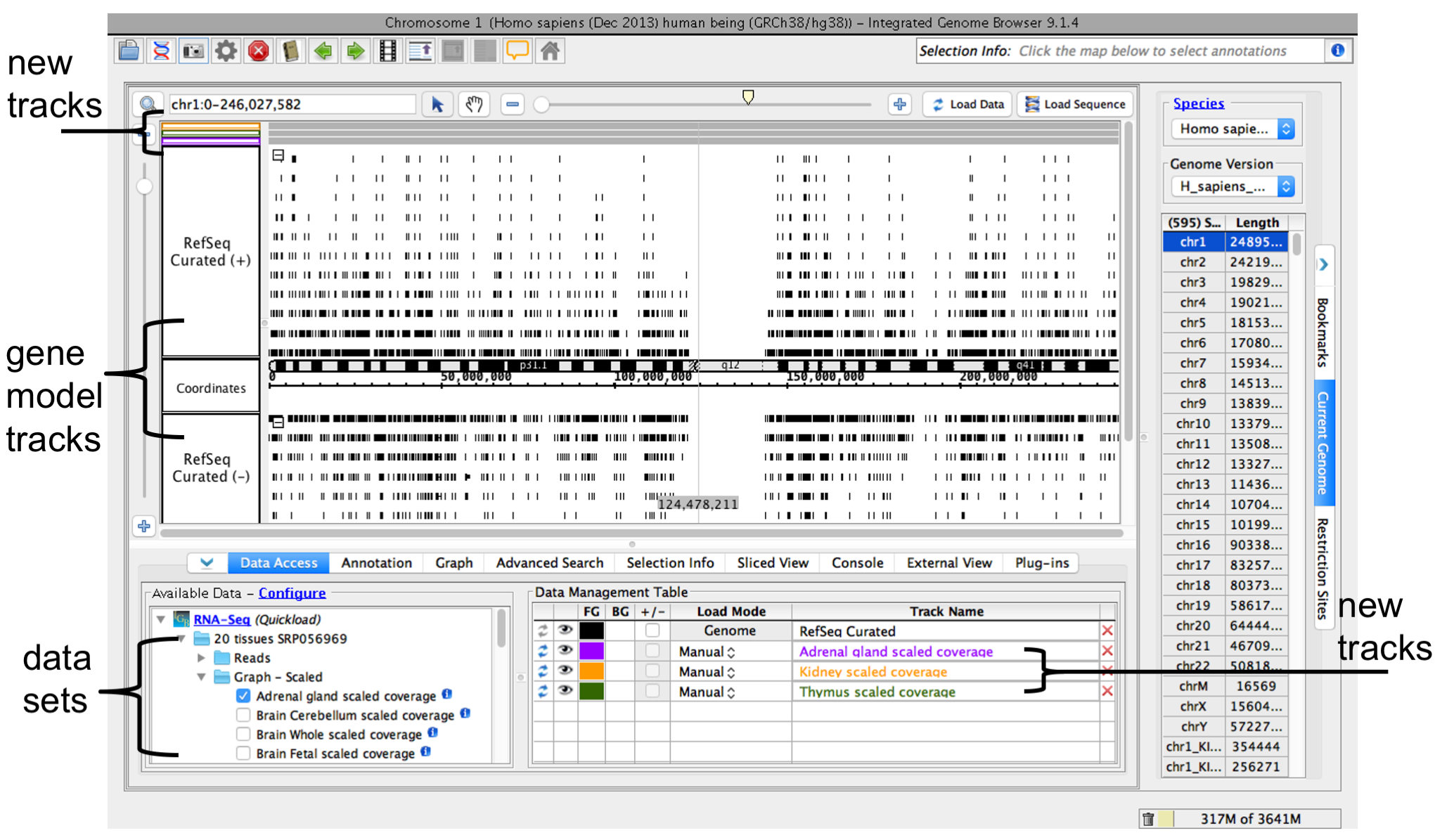
Open data sets from remote data sources (Data Access tab) or by opening local files.
To open a data set from a remote an IGB Quickload data source (see About IGB Quickload)
- Select Data Access
- Select data sets in the Available Data section
...
When you select a data sets or a file, IGB will add adds a new empty track to the main view and list lists it in the Data Management table. Regions Empty regions in the new track that do not have data loaded have a darker background colorare gray.
IGB window after opening a data sethuman genome RNA-Seq coverage graphs from Adrenal Gland, Kidney, and Thymus data sets
Step 4: Zoom in
...
IGB horizontal zooming controls
Other ways to zoom
...
- Search for a gene by name or keyword (For example, TBATA or thymus)
- Double-click a featurean exon or gene model to zoom in on it
- Click-drag within the coordinate axis to zoom in on a region
See also:
...
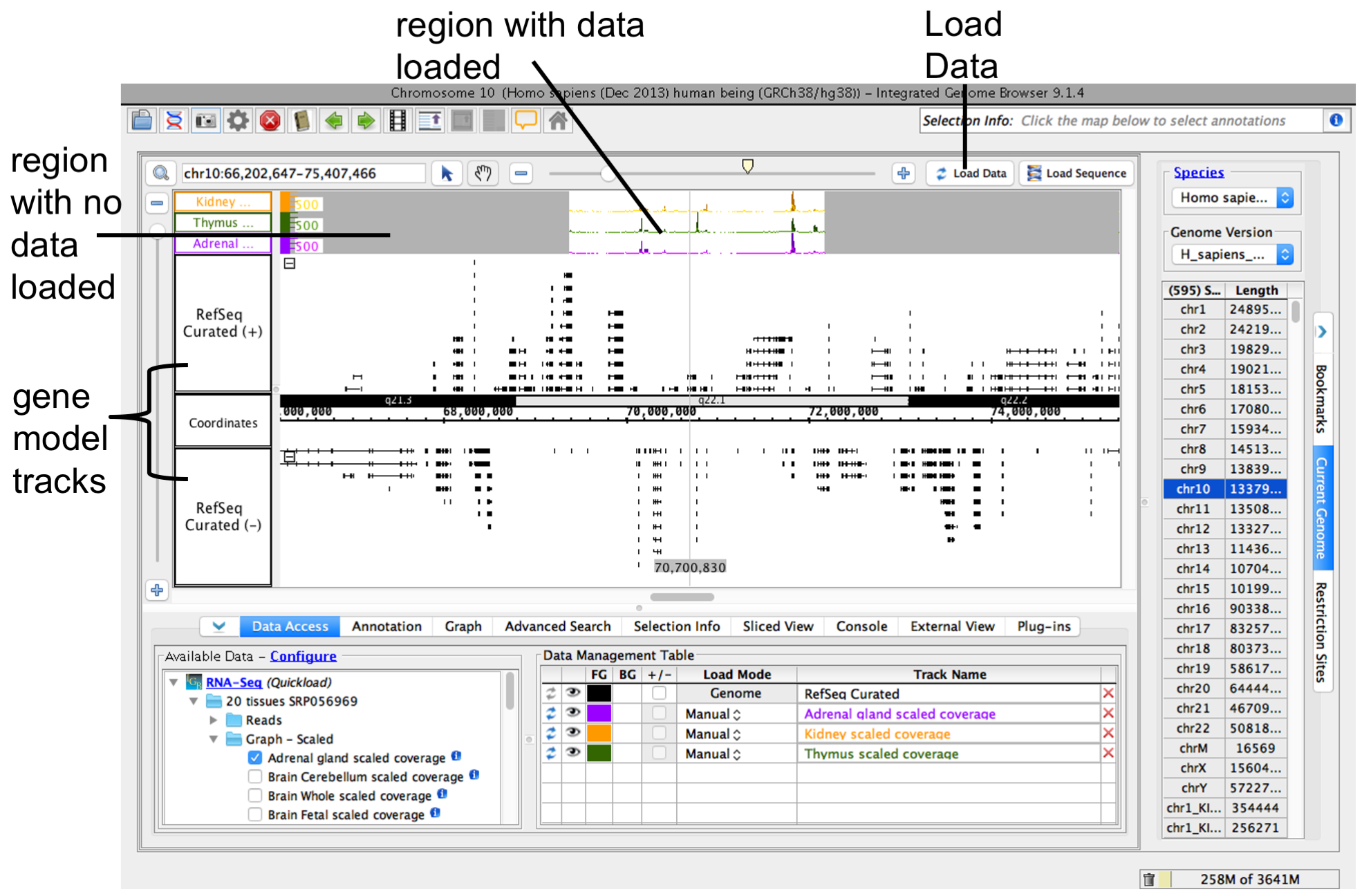
To load data, click Load Data button. Regions with loaded data show the selected background color; areas without loaded data appear darker.
IGB after loading data
See also:
...