Introduction
Graphs contain Genome graphs associate numerical data associated with base pair positions along the chromosome sequence axis, such as sets of scores or chromosome copy numbers at specific positions. IGB allows you to visualize such graph data and compare it with RefSeq and other annotations.
Examples of suitable data include:
...
. You can open and view genome graph data files in IGB and manipulate them using functions in the Graph tab.
Using the Graph tab settings, you can display graphs as line graphs or heat graphs, change their scale, convert them to floating (draggable) graphs, and perform mathematical or other types types of transformations. You can also compute averages or sums of multiple graphs.
You can create graphs from annotation tracks, such as an RNA-Seq coverage graphs showing the number of read alignments starting at each position.
Examples of genome graphs include:
- Probe intensity values from genome tiling arrays.
- Density of ESTs across Percentage methylation across chromosomes
- GC content along chromosomes
- Measures of conservation between two genomes
There are two basic types of genomes graphs:
- Position graphs associate scores with single genomic positions.
- Interval graphs associate scores with ranges of genomic positions.
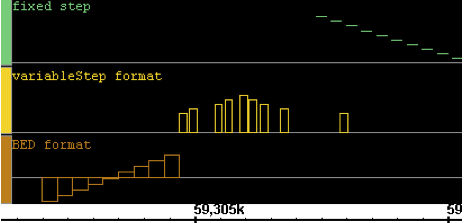
Here is a very simple example of several interval graphs:
IGB will recognize whether the graph is a position or interval graph and will display it accordingly.
This chapter explains how to use IGB to examine your graph data:
- Compare graph and annotation data in the same region.
- Adjust graphs to highlight features
- Fine-tune graph scaling to improve visibility
*
Use thresholding to transform graph data into annotation-like format, in order to examine it further using other tools in IGB.positions