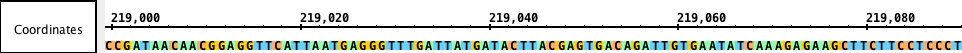| Table of Contents |
|---|
...
Genomes and
...
chromosomes
chromosome: In IGB, chromosome refers to any single sequence. Typically this corresponds to the sequence of a physical chromosome. Alternatively, it may represent an assembled contig corresponding to part of a physical chromsome. All chromosomes in IGB are assumed to be DNA, rather than RNA sequences. No two chromosomes within the same genome version can have the same name.
...
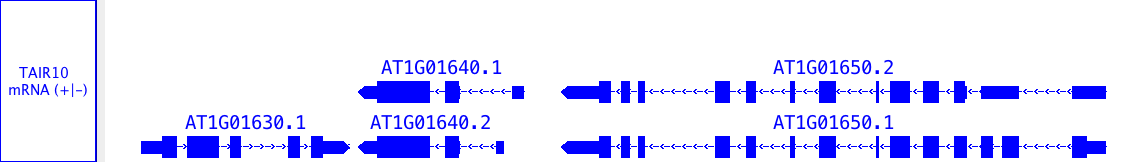
- single-coordinate feature: splice sites
- single-span feature, with strand: an exon
- single-span feature, no strand: a restriction site
- multi-span feature, with strand: a gene model
...
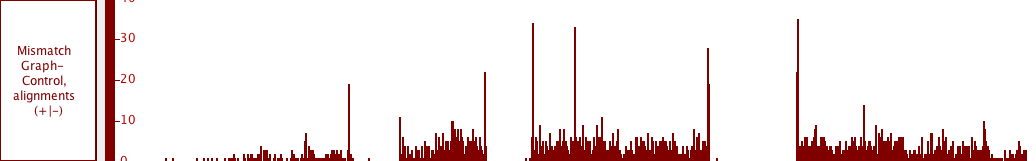
Graphs
Graphs consists of are numeric data associated with regions or single-base positions in a chromosome.
...
Sequences
Sequences are sets of DNA residues comprising a chromosomefrom a chromosomes or contigs that together make an assembly. Sequences can be fully or partially loaded from local files, QuickLoad Quickload sites, or DAS servers. IGB allows you to examine sequence data in a separate window called Distributed Annotation Servers. You can view sequence data in the Coordinates track or by opening the Sequence Viewer.
...
Alignments
Alignments represent how sequences obtained from an experiment (such as reads from an RNA-Seq experiment) align onto the a reference genomic sequence. At low zoom they look like annotations, but with marks representing mismatches between the read and the reference genome whenever these data are available, insertions, or deletions. At higher zoom, the aligned sequence bases become visible. File formats for alignments include BAM and PSLX.
...
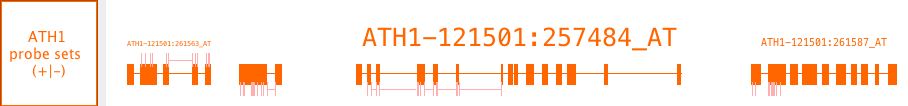
Probe set alignments
Probe set alignments consist of a probe set target sequence sequences aligned onto a chromosome and the location of probes relative to the aligned target sequence. This data type is Affymetrix-the reference with probe locations indicated as annotations on the target sequence alignments. The probes are kind of annotation on an annotation.
These data are Affymetrix specific.