IGB can integrate and merge data from multiple sources in the same view. These includes include data sets loaded from your computer, from UCSC Genome Browser, from URL sites, or from various public (and private) DAS, DAS2, and Quickload servers.
To load data:
| Table of Contents |
|---|
...
Choose Species and Genome Version
Use the Current Genome tab to select a Species and Genome Version. Or , or select an image on the Start Screen.
...
Once you select a species and genome version, publicly accessible data sets hosted on by UCSC Genome Browser, IGB Quickload sites or , and other sources will appear in the Available Data section of the Data Access tab. Select these data sets to open them in IGB.
You can also open files in IGB if they're stored locally or via a publicly-available URL. To open a file, select File > Open File... or File > Open URL...
...
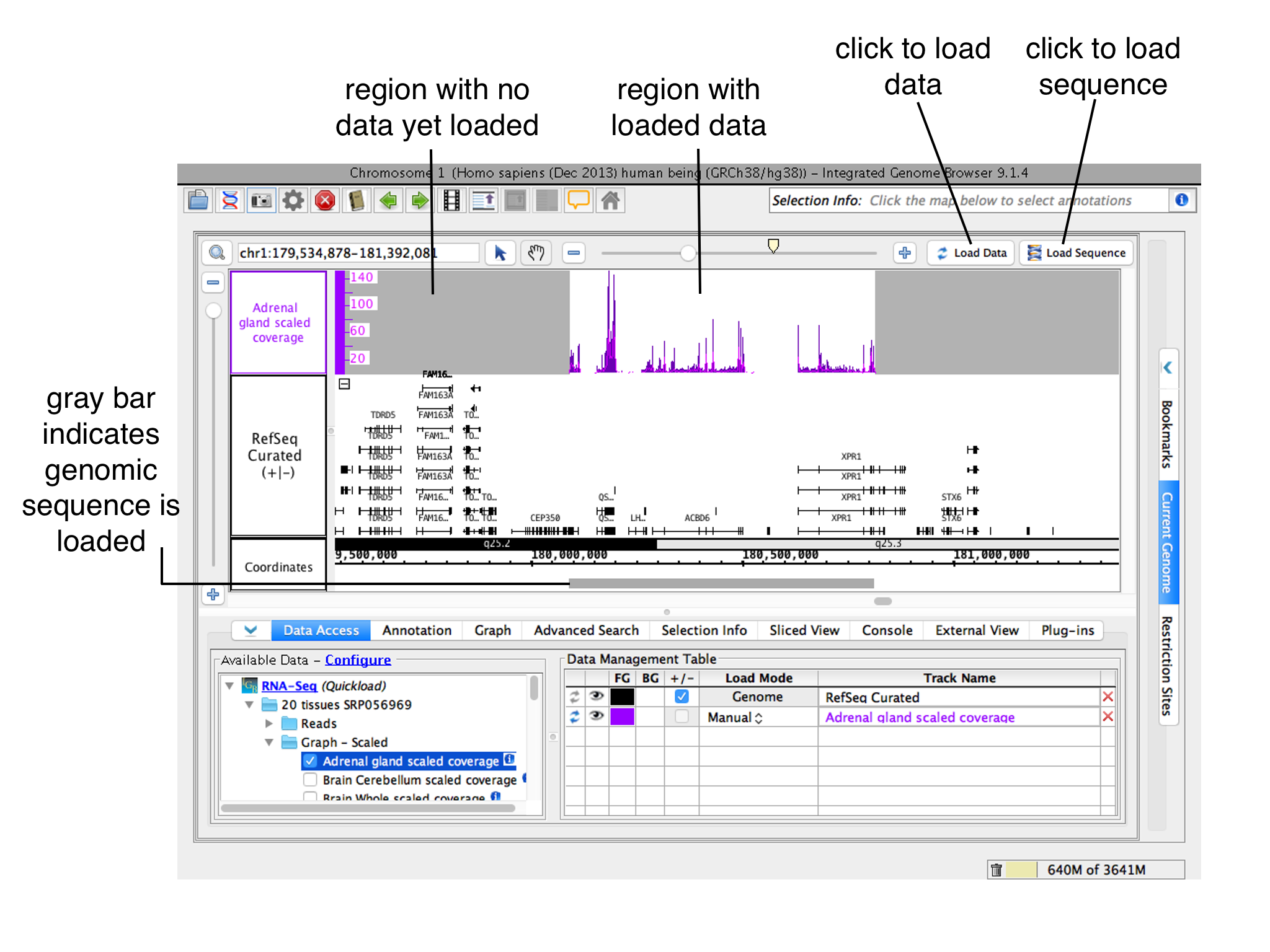
To change how and when IGB loads data into a track, open the Data Access panel and change settings under Load Mode in the Data Management Table.
Options include:
- Don't Load means no data will load when you click the Load Data button.
- Genome triggers automatic loading of all data from a file.
- Manual means that only data overlapping for the currently displayed region will load when you click Load Data. This is the default setting.
- Auto means that moving or zooming to a new location triggers data loading, but only when the zoom threshold is lower than the mark shown on the horizpontal horizontal zoomer. Change the threshold by selecting selecting View > Set AutoLoad Threshold to Current View.
...
Regions where data have not yet been loaded have gray background color.