...
- Add a Data Provider to IGB using the following URL: http://igbquickload.org/smokeTestingQuickload/
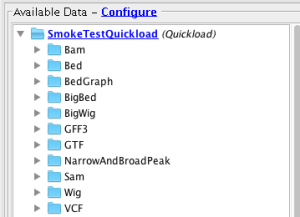
- The added Quickload site is available for the Homo sapien genome under the Available Data pane.
- Verify that all files in the Quickload site are loading into IGB:
- Bam
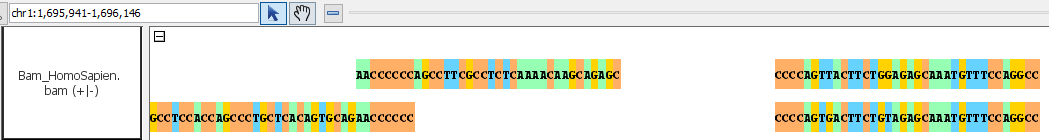
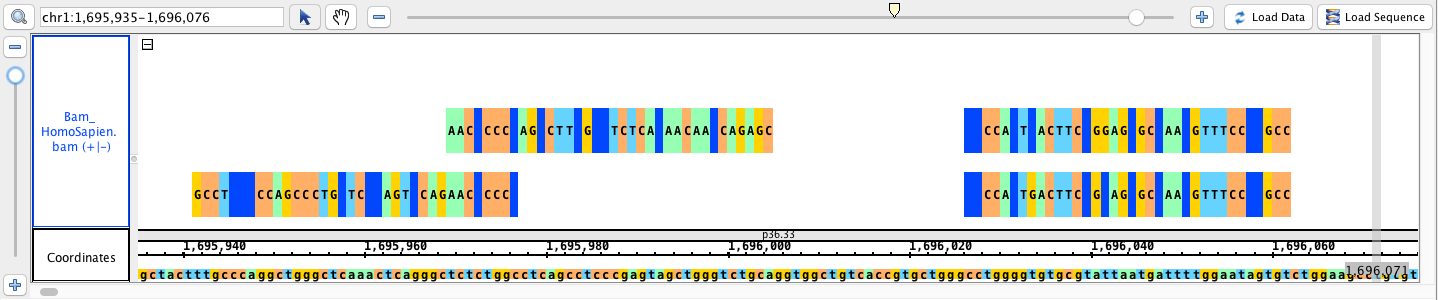
- Bam_HomoSapien.bam
bam
go to region: chr1:1,695,935-1,696,076
To get the dark blue blocks you will need to load the sequence.
- Bam_HomoSapien.bam
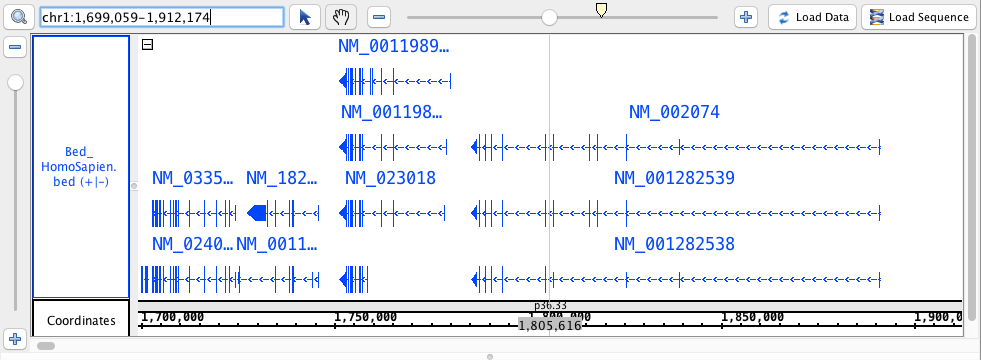
- Bed
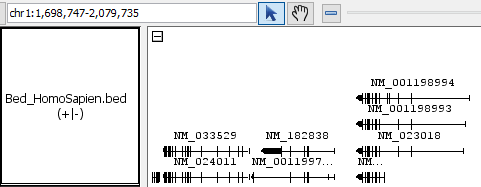
- Bed_HomoSapien.bed
- Bed_HomoSapien.bed.gz
region: chr1:1,699,059-1,912,174
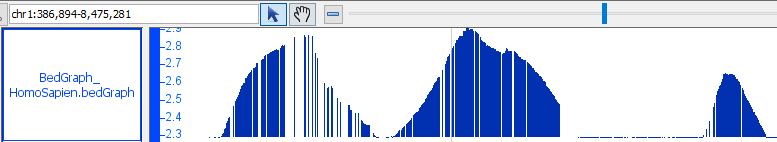
- BedGraph
- BedGraph_HomoSapien.bedgraph
- BedGraph_HomoSapien.bedgraph.gz
chr1:386,893-7,895,983
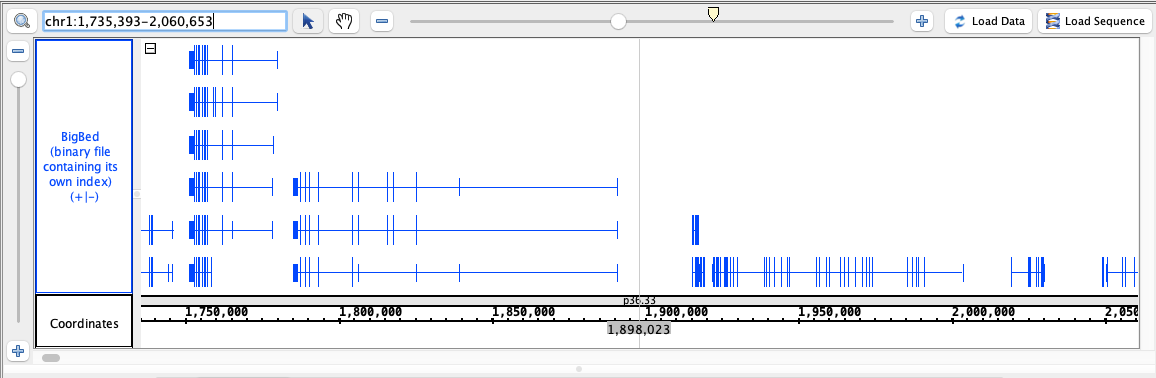
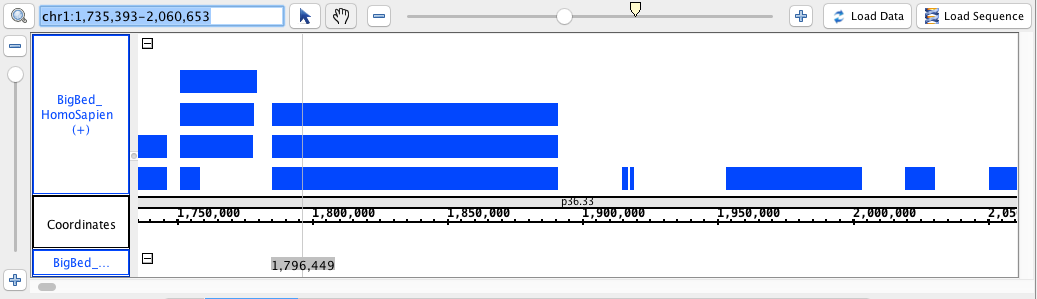
- BigBed
- BigBed_HomoSapien.bigbed
chr1:1,735,393-2,060,653
- BigBed_HomoSapien.bigbed
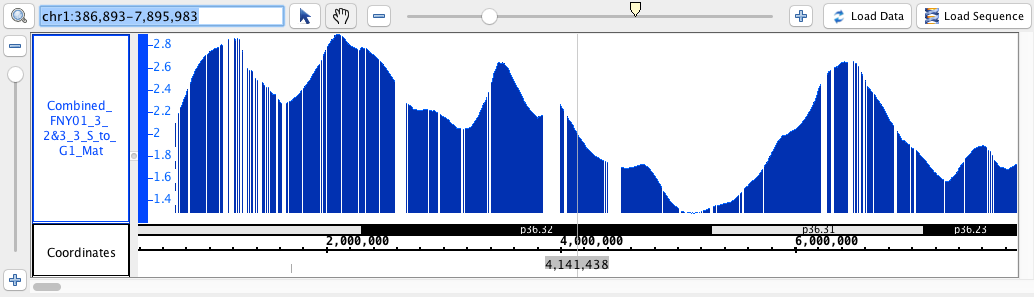
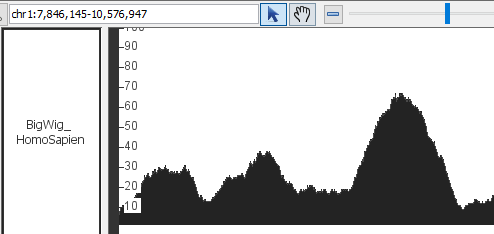
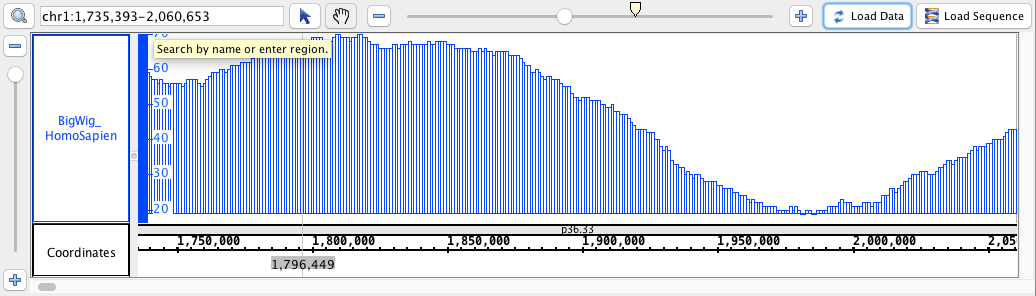
- BigWig
- BigWig_HomoSapien,bigwig
chr1:1,735,393-2,060,653
- BigWig_HomoSapien,bigwig
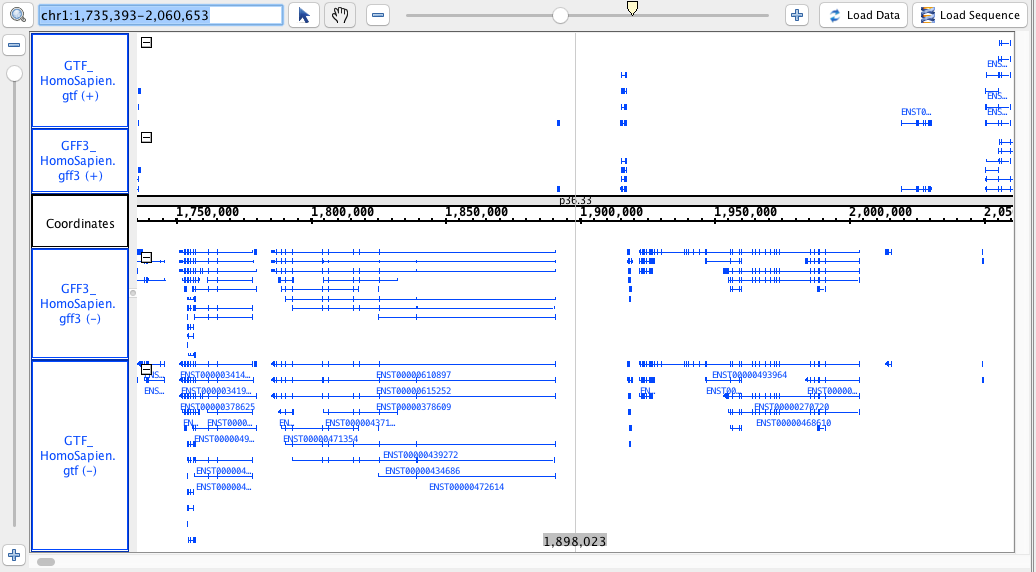
- GFF3
- GFF3_HomoSapien.gff3
- GFF3_HomoSapien.gff3.gz
- GTF
- GTF_HomoSapien.gtf
- GTF_HomoSapien.gtf.gz
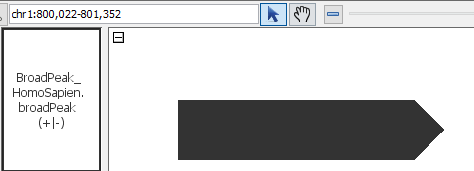
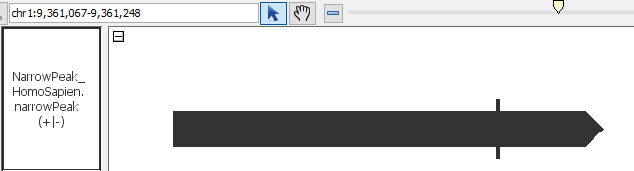
- NarrowAndBroadPeak
- BroadPeak_HomoSapien.broadpeak
- BroadPeak_HomoSapien.broadpeak.gz
- NarrowPeak_HomoSapien.narrowpeak
- NarrowPeak_HomoSapien.narrowpeak.gz
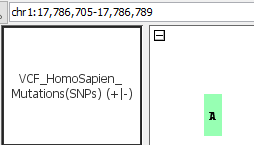
- VCF_HomoSapien_Mutations(SNPs).vcf
- VCF_HomoSapien_Mutations(SNPs).vcf.gz
- Can you see mutations (SNPs)?
- Can you see mutations (SNPs)?
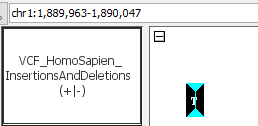
- VCF_HomoSapien_InsertionsAndDeletions.vcf
- chr1:0-12,643,524
- VCF
- VCF_HomoSapien_InsertionsAndDeletions.vcf.gz
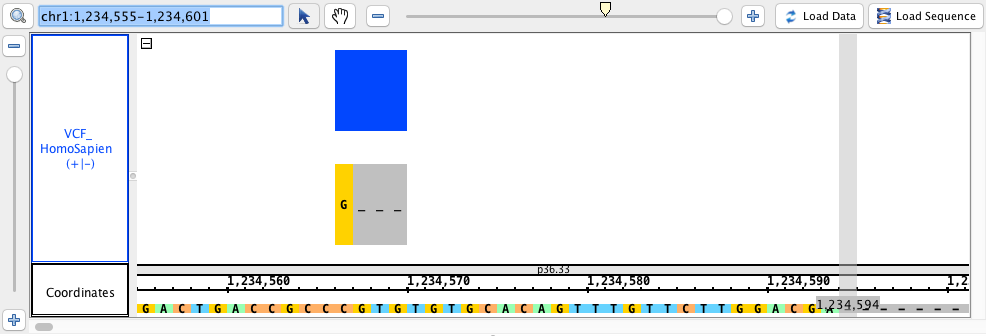
- Can you see deletions?
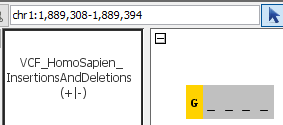
- Can you see insertions?
chr1:1,234,555-1,234,601 - Can you see deletions?
- VCF_HomoSapien_InsertionsAndDeletions.vcf.gz
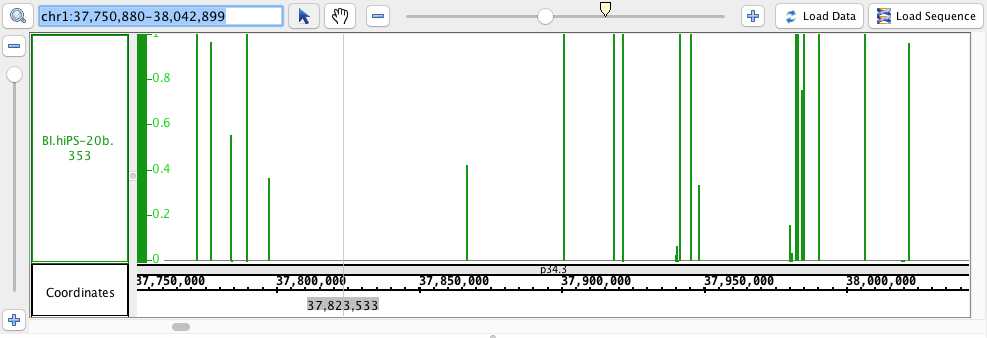
- Wig
- Wig_HomoSapien.wig
- Wig_HomoSapien.wig.gz
chr1:37,750,880-38,042,899
- Bam
- The added Quickload site is available for the Homo sapien genome under the Available Data pane.
- Add a Data Provider to IGB using the following URL: http://igbquickload.org/smokeTestingQuickload/
Sequence appearance
Go to the Arabidopsis genome. From the RNA-Seq quicklaod (available by default as of 9.0.1), open the file RNA-Seq / Pollen SRP022162 / Reads / Pollen alignments. (url http://lorainelab-quickload.scidas.org/rnaseq/A_thaliana_Jun_2009/SRP022162/Pollen.bam)
...