General Function Checklist
- Protannot can ProtAnnot is be launched from IGB
- Go to
- Install the ProtAnnot plugin through the App Manager (Tools>Open App Manager)
- Select any genome
- Select one or more whole gene model annotation
- Select the Tools Menu
- Select Start ProtAnnot
- Checkpoint
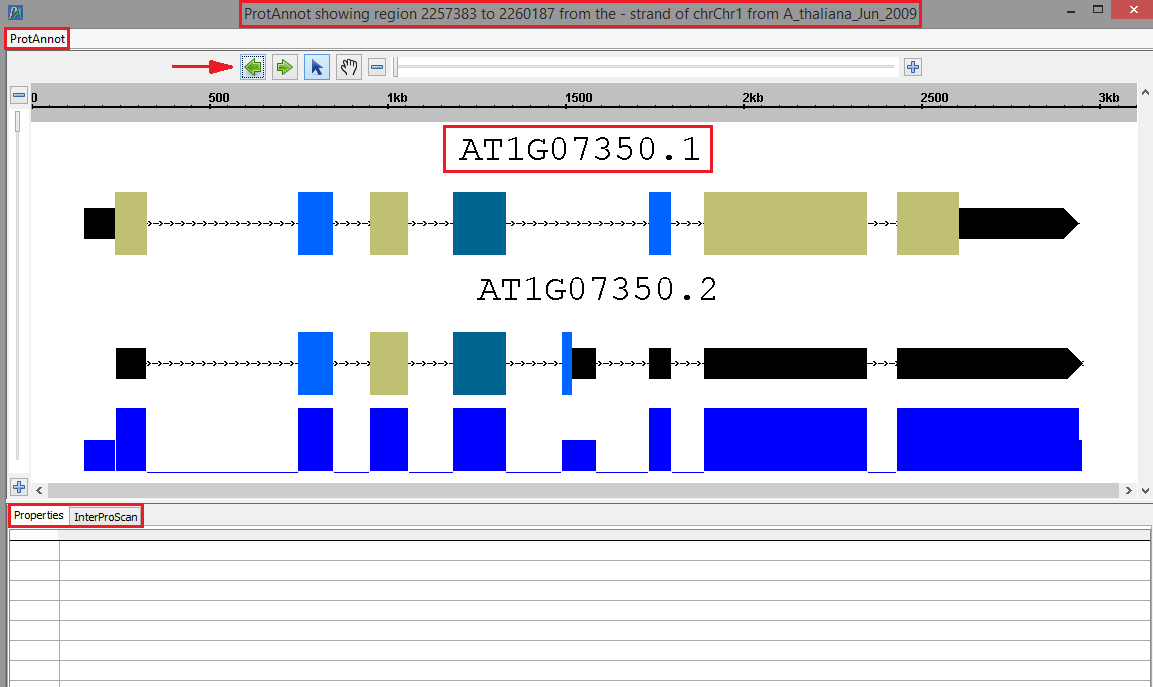
- sr45a Your selected gene model(s) should look as follows when loaded into ProtAnnot. Take ; take note of the following key items in red.
...
- :
- The window title
- The ProtAnnot menu
- The navigation and zoom bar
- Th selected gene model(s) name
- The Properties and InterProScan tabs
- :
- The ProtAnnot window title should contain the absolute sequence coordinates, strand direction, contig name, and the genome version.
- InterProScan
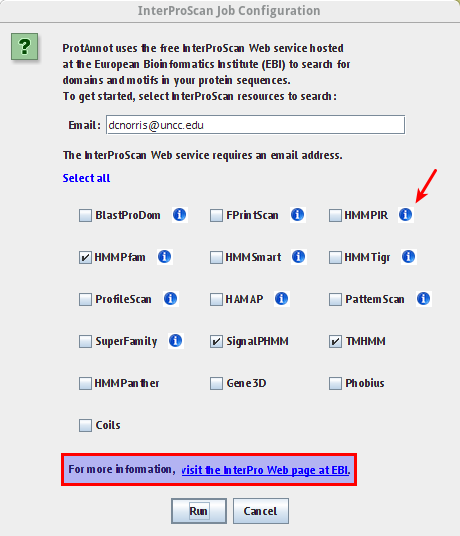
- Select ProtAnnot menu
- Select Load InterProScan... menu item
- Verify the InterProScan Job Configuration window appears similar to the image below
- Verify tool tips in the information icon wrap appropriately
- Verify the link in the highlighted box below links to the appropriate InterProScan about page
- Select run
- {{ image of InterProScan results }}
- Verify that the Exons have the correct protein sequences
...
- Run InterProScan again by selection "Run InterProScan" under the InterProScan tab. See image above.
- Once the jobs are running select "Cancel All Jobs" under the InterProScan tab
- Verify that the job statuses are "Cancelled" and that there are no exceptions in the logs.
o Your selected gene model(s) should look as follows when loaded into ProtAnnot. Take note of the following key items in red:
§ The Window Title
§ The ProtAnnot Menu
§ The navigation and zoom bar
§ The selected gene model(s) name
§ The Properties and InterProScan tabs